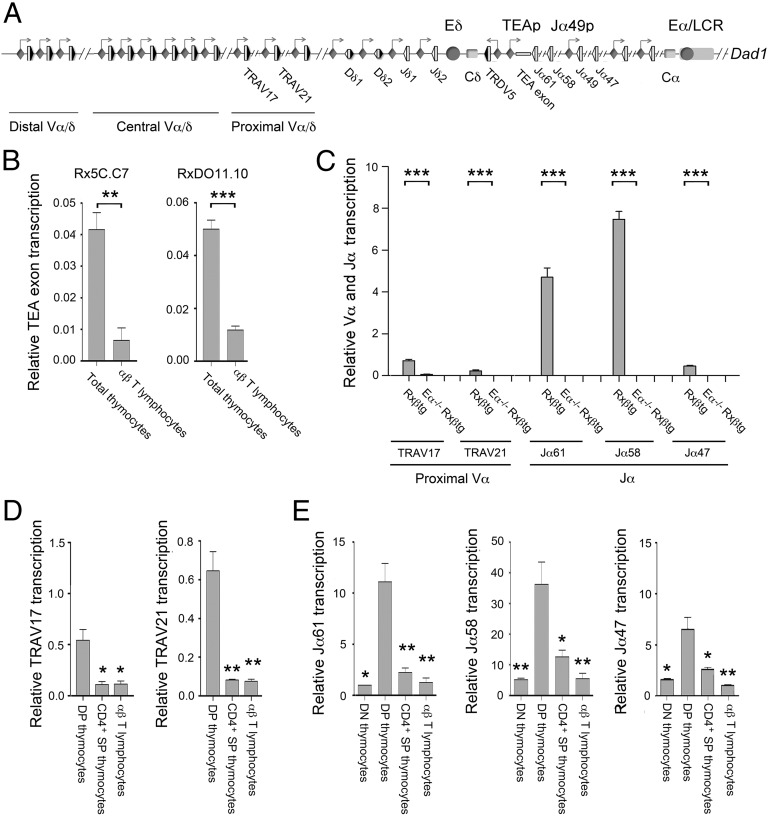
Fig. 1.
Tcra Eα-dependent transcription is inactivated in SP thymocytes and αβ T lymphocytes. (A) Schematic representation of Tcra/Tcrd genomic structure. The V, D, and J gene segments are represented by gray vertical rectangles, and the recombination signal sequences are shown as black (containing a 23-bp spacer) or white (containing a 12-bp spacer) triangles. Positions of the TEA exon, the Vα/δ, Dδ, Jδ, and Jα gene segments, and the Cδ and Cα regions are indicated. Arrows represent active sites for germ-line transcription. Promoters and enhancers are represented as diamonds and circles, respectively. The Tcra LCR is represented as a long horizontal rectangle. (B) Analysis of TEA exon transcription in Rx5C.C7 and RxDO11.10 thymocytes and T lymphocytes by RT-qPCR. (C) Analysis of proximal Vα and Jα transcription in DP thymocytes from Rxßtg and Eα−/− Rxßtg mice by RT-qPCR. (D) Analysis of TRAV17 and TRAV21 germ-line transcripts in RxDO11.10 sorted thymocyte populations and T lymphocytes by RT-qPCR. (E) Analysis of Jα transcription in RxDO11.10 sorted thymocyte populations and T lymphocytes by RT-qPCR. The results were normalized to those for Actb and represent the mean ± SEM of duplicate RT-qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the values of the RT-qPCRs from total thymocytes vs. T lymphocytes (B), Rxßtg vs. Eα−/− Rxßtg thymocytes (C), and DP thymocytes vs. T lymphocytes (D and E) (*P < 0.05, **P < 0.005, and *** P < 0.0005).