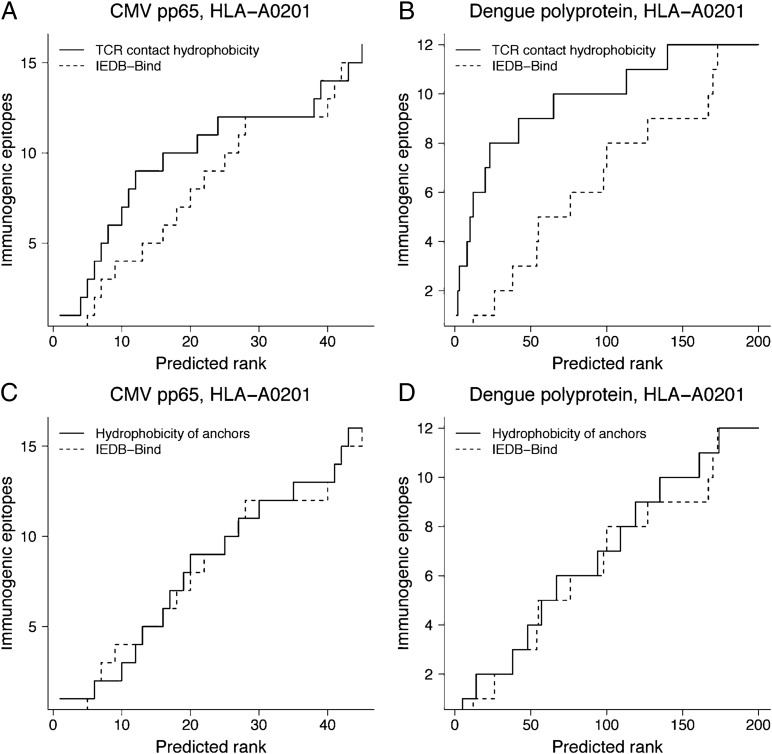
Fig. 3.
Efficiency of predicting experimentally defined HLA-A0201–restricted immunogenic epitopes using mean hydrophobicity of TCR contact residues (straight lines) compared with IEDB consensus binding tool (IEDB-Bind; dashed lines). Tegument protein pp65 from cytomegalovirus (CMV) and polyprotein from dengue virus type 1 were used for predictions. (A and B) Predicted peptides from the IEDB-Bind were reranked using the mean hydrophobicity of TCR contact residues. (C and D) Predicted peptides from the IEDB-Bind were reranked using the mean hydrophobicity of anchor residues.