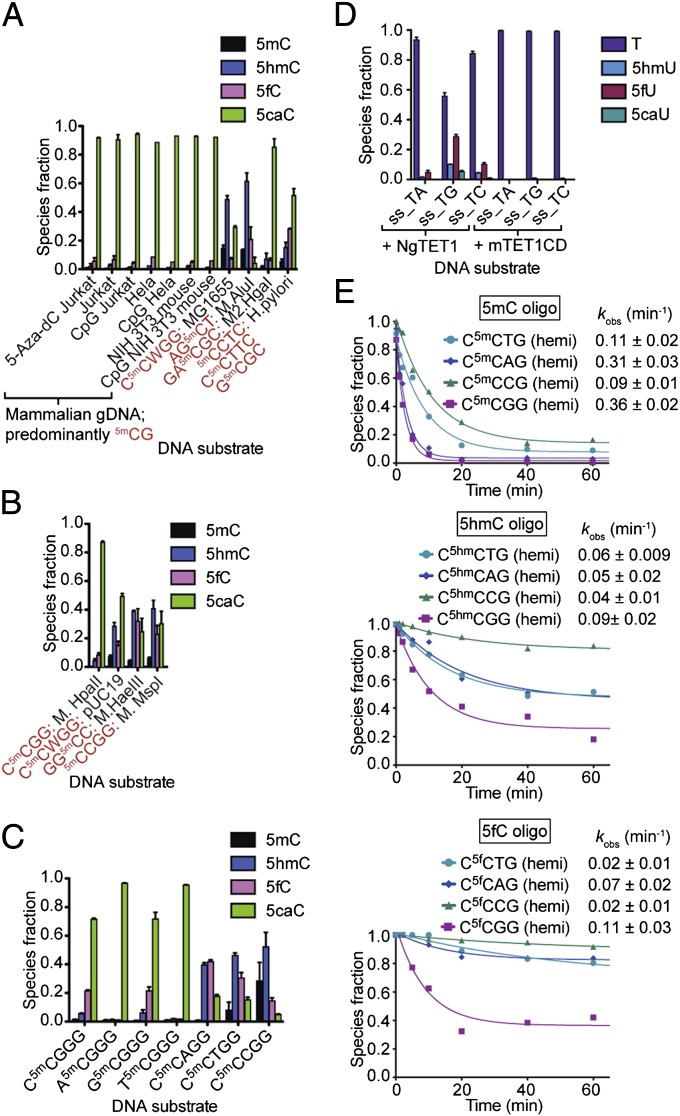
Fig. 3.
NgTET1 activity is dependent on nucleotide-sequence context. Distribution of NgTET1 reaction species: (A and B) As quantified by LC-MS (Agilent 1200), using excess enzyme (20 µM) with (A) genomic (2.5 μg, sheared to 1.5-kb) or (B) plasmid (2.5 μg) DNA containing different methylation sequences as indicated in red (Table S2); (C and D) As quantified by 6490 Triple Quad LC-MS for (C) 30-min reaction of 8 μM NgTET1 with 4 μM oligo DNA or (D) 6.7 μM NgTET1 (in Mops buffer pH 6.9) or mTET1CD with 1.6 μM oligo DNA. For A–D, error bars (in black) represent the SEM (n ≥ 3). (E) Kinetic traces, with species fraction determined by LC-MS (Agilent 1200), of NgTET1 with 5mC-, 5hmC-, or 5fC-containing oligos hemimethylated at a single CpX site (Table S1). Reactions were done in Mops buffer (pH 6.75) using 8 μM NgTET1 and 4 μM DNA. The data are fit to a single exponential and the observed rate constants with SEM are provided.