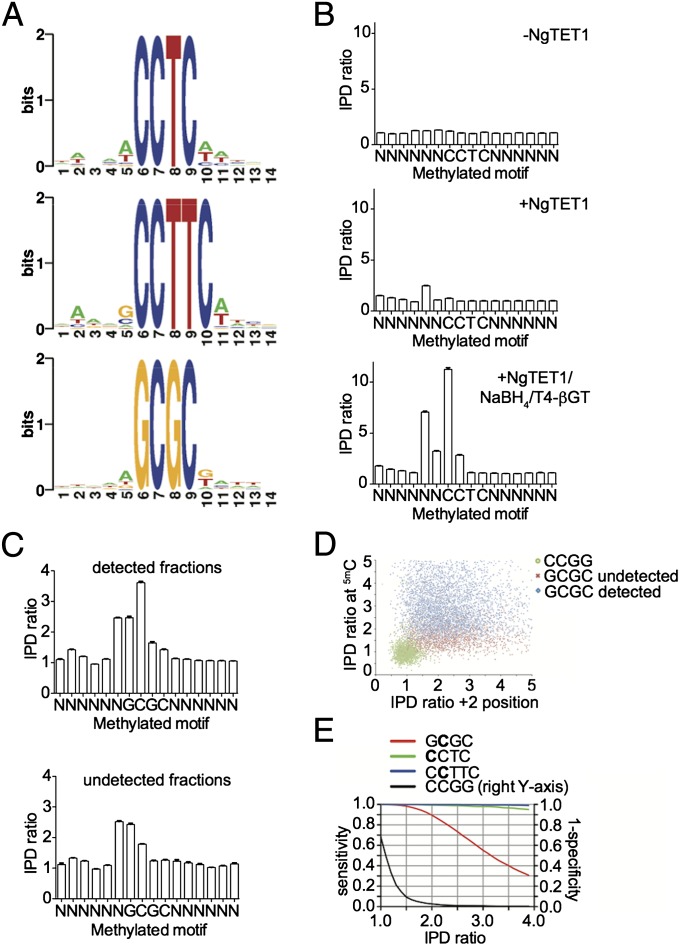
Fig. 4.
SMRT sequencing of H. pylori gDNA using NgTET1. (A) Sequence logos for 5mCCTC, 5mCCTTC and G5mCGC motifs detected by SMRT sequencing of NgTET1-treated gDNA. (B) IPD ratio plots corresponding to the 5mCCTC motif in gDNA treated in the absence of NgTET1 (Top), with NgTET1 (Middle), or with NgTET1/NaBH4/T4-βGT (Bottom). (C) IPD ratio plots for the sequences detected (Upper) versus undetected (Lower) as belonging to the G5mCGC motif for gDNA treated with NgTET1/NaBH4/T4-βGT. For B and C, the error bars (in black) represent the SEM. (D) Scatter plot of IPD ratio values at the methylated and +2 positions for G5mCGC and CCGG sequences for gDNA treated with NgTET1/NaBH4/T4-βGT. (E) Plot of sensitivity and specificity as a function of IPD ratio for gDNA treated with NgTET1/NaBH4/T4-βGT.