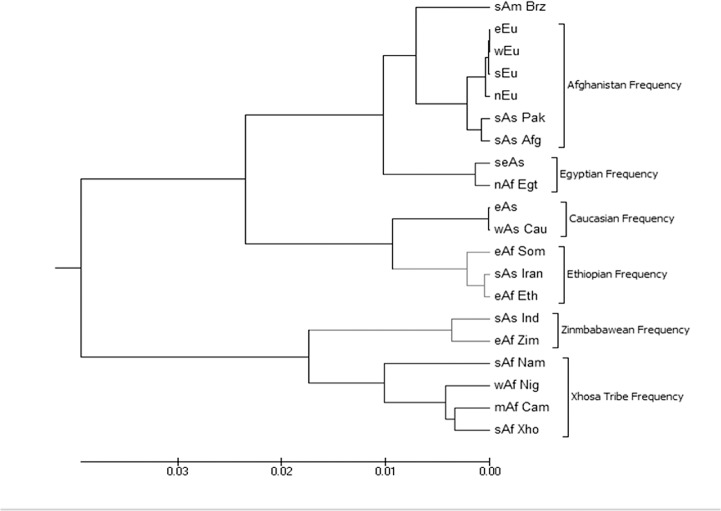
Fig 1. Phylogenetic tree of 20 different continental regions population affiliations for GST M1-T1 null allele frequency.
The tree was produced by the UPGMA method from DST values in Table 2 and cluster with more than 50% of 1000 bootstrap replicates were included in the consensus tree obtained by Felsenstein (1989) phylogeny interference package. Major group of GST M1-T1 null allele frequencies were from population of Xhosa tribe, Zimbabwe, Ethiopia, Egypt, Afghanistan and Caucasian. Abbreviations used were same as those in Table 1.