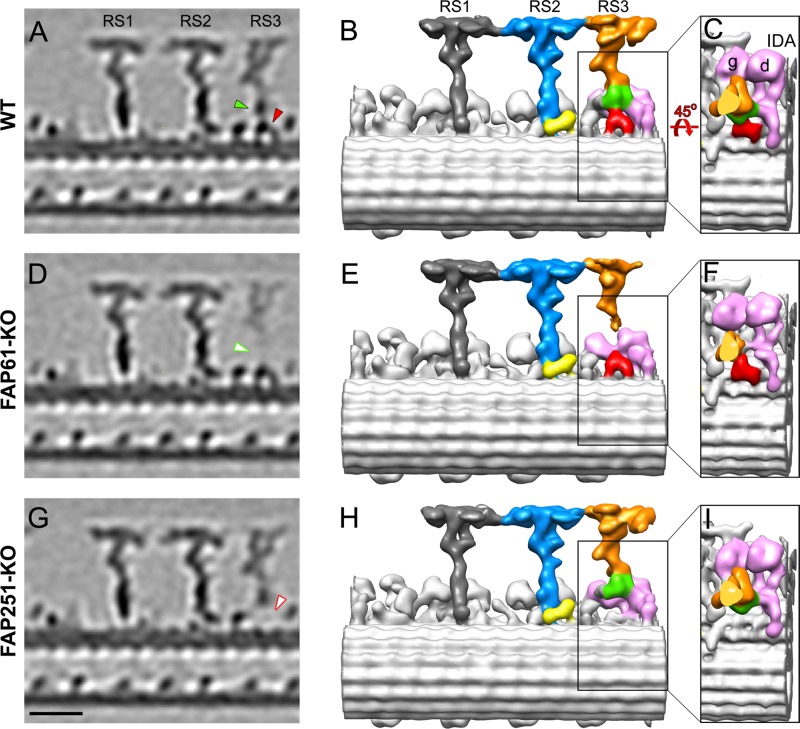
FIGURE 3:
Comparison of the 3D structures of the 96-nm repeats from Tetrahymena WT, FAP61-KO, and FAP251-KO axonemes. Tomographic slices (A, D, G) and isosurface renderings (B, C, E, F, H, I) show the averaged 96-nm repeats in longitudinal views in WT (A–C) and the two CSC-knockout mutants, FAP61-KO (D–F) and FAP251-KO (G–I). In the WT strain, the central stem region of RS3 (green) connects to the arch-like basal region (red), the main distal part of RS3 (orange), and the tails of the two dynein IDAs g and d (pink; B, C). Both CSC-knockout mutants show structural defects in the basal half of RS3: in FAP61-KO (D, E), the central stem region (green arrowhead in A), and in FAP251-KO (G, H), the base (red arrowhead in A) are barely visible. Note that the main distal part of RS3 (orange) is present in all strains, but in FAP61-KO, its electron microscopic density is significantly weaker (D). The other two radial spokes, RS1 (dark gray) and RS2 (blue/yellow), appear structurally normal in the mutants compared with WT; note that also the back prong of RS2 (yellow), which was reduced in the artificial microRNAi mutants of FAP61 and FAP91 in Chlamydomonas (Heuser et al., 2012), seems unaffected in the Tetrahymena FAP61-KO and FAP251-KO mutants studied here. Bar, 20 nm.