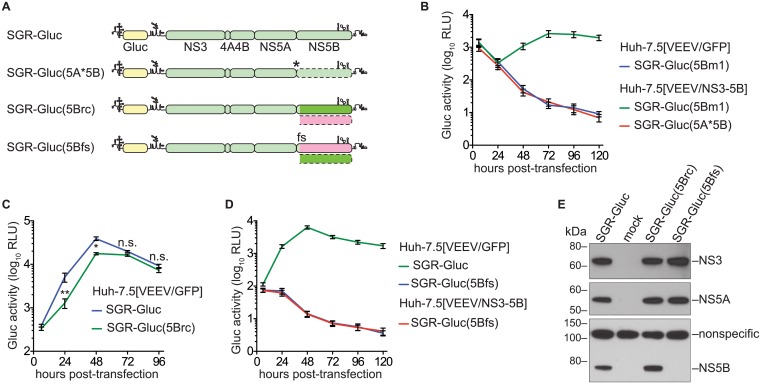
Fig 2. NS5B protein is required in cis.
(A) Constructs used to examine the cis-requirement for NS5B expression, as detailed in the text; asterisk, stop codon; dotted lines, untranslated coding regions; fs, -1 frame-shift. For more information, also see Table 1 and S2 Text. (B) SGR-Gluc(5A*5B) does not replicate and is not complemented in trans. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls. This experiment was performed three times with similar results. (C) SGR-Gluc(5Brc) replicates efficiently. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls; n.s., p ≥ 0.05; *, p < 0.05; **, p < 0.01 by Students t-test. This experiment was performed three times with similar results. (D) SGR-Gluc(5Bfs) does not replicate and is not replicated in trans. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls. This experiment was performed three times with similar results. (E) The SGR-Gluc(5Brc) and SGR-Gluc(5Bfs) polyproteins are expressed and properly cleaved, as determined by western blot with 4D11 anti-NS3 and 9E10 anti-NS5A monoclonal antibodies. Because these mutants do not replicate, expression was driven by the vaccinia-T7 RNA polymerase system.