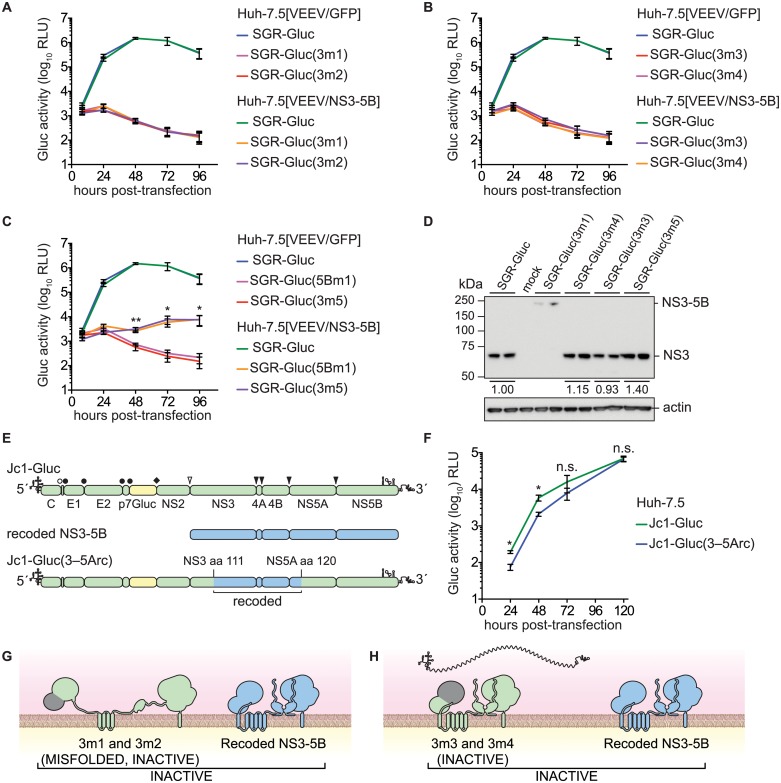
Fig 3. Complementation of NS3 serine protease and RNA helicase active site mutants.
(A) Serine protease active site mutants SGR-Gluc(3m1) and SGR-Gluc(3m2) do not replicate and are not complemented in trans. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls. This experiment was performed two times with similar results. (B) Helicase RNA binding and NTPase active site mutants SGR-Gluc(3m3) and SGR-Gluc(3m4) do not replicate and are not complemented in trans. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls. For comparison, the SGR-Gluc samples, which were performed in parallel, are reproduced from panel A. This experiment was performed two times with similar results. (C) RNA helicase base stacking mutant SGR-Gluc(3m5) does not replicate but is complemented in trans with an efficiency similar to SGR-Gluc(5m1). For comparison, the SGR-Gluc samples, which were performed in parallel, are reproduced from panel A. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls; *, p < 0.05; **, p <0.01 by Student’s t-test, comparing matched time points from SGR-Gluc(3m5) in Huh-7.5[VEEV/NS3–5B] vs. Huh-7.5[VEEV/GFP] cells. This experiment was performed two times with similar results. (D) Western blot to confirm that the serine protease active site mutant SGR-Gluc(3m1) does not undergo polyprotein cleavage and that the RNA helicase mutants SGR-Gluc(3m2), SGR-Gluc(3m3), SGR-Gluc(3m4), and SGR-Gluc(3m5) do not exhibit defects in NS3 accumulation. NS3 was detected by using anti-NS3 monoclonal antibody 4E11; values represent relative NS3 levels compared to SGR-Gluc. Because these mutants do not replicate, expression was driven by the vaccinia-T7 RNA polymerase system. (E) Design of the Jc1-Gluc(3–5Arc) mutant. (F) Jc1-Gluc(3–5Arc) replicates efficiently. Values represent mean ± SD from transfections done in triplicate and normalized to untransfected controls; n.s., p >0.05, *, p ≤ 0.05 by Student’s t-test. This experiment was performed two times with similar results (G) Model showing that SGR-Gluc(3m1) and SGR-Gluc(3m2) do not undergo proteolytic processing, produce misfolded, inactive polyproteins, and are not complemented in trans; grey signifies the nonfunctional serine protease domains; blue signifies the recoded NS3-5B polyprotein. (H) Model showing that SGR-Gluc(3m3) and SGR-Gluc(3m4) are not trans-complemented and may have a defect in RNA recruitment (compare to Fig 1B); grey signifies the nonfunctional helicase domains; blue signifies the recoded NS3-5B polyprotein.