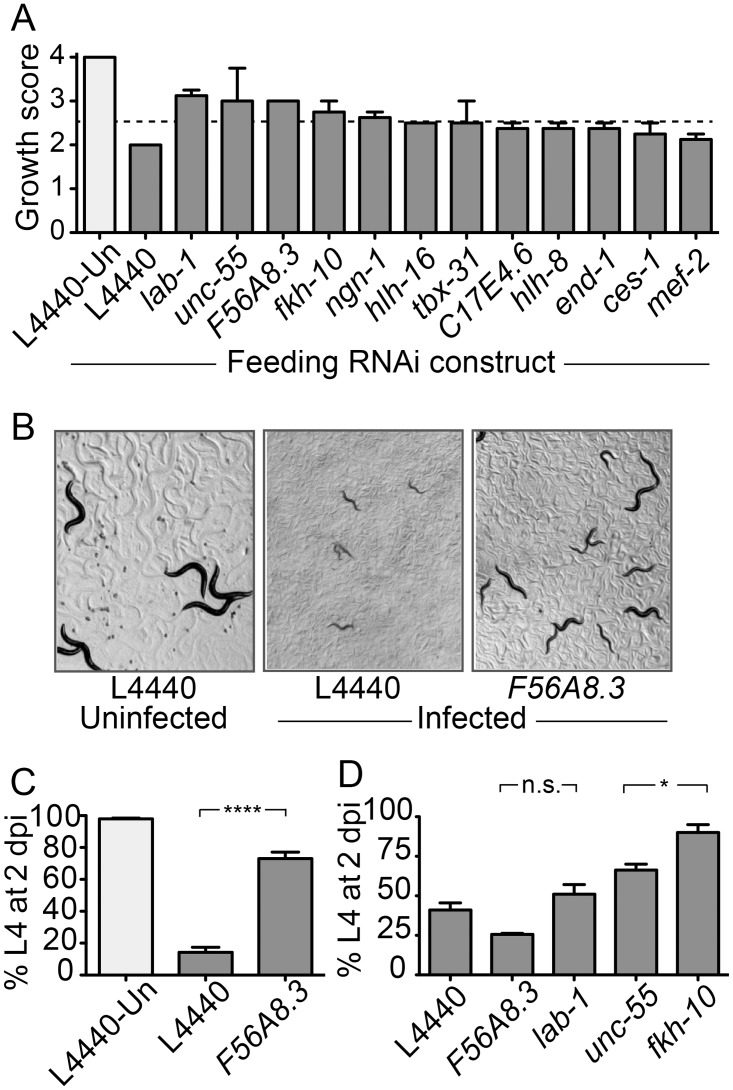
Fig 1. RNAi screen for host genes that regulate N. parisii-induced larval arrest of C. elegans.
(A) Twelve RNAi clones that were originally scored as hits in the screen were retested, using a semi-quantitative visual score of C. elegans eri-1 strain growth on feeding RNAi at 2 days post-infection (dpi). Uninfected animals grown on L4440 (control RNAi, light gray) provided the baseline for fully developed worms and were scored as a four, while fully arrested worms were scored as a one. Re-tested RNAi clones were scored under infected conditions (dark gray). Twelve hits are shown, consisting of 10 transcription factors, one LRR gene (F56A8.3), and one contaminant (gene identity different from what was listed in the library) belonging to neither gene class (lab-1). Data are represented as mean values with standard error of the mean (SEM) from biological duplicates in two independent screens. (B) Representative images of larval arrest upon N. parisii infection, and inhibition of arrest after F56A8.3 RNAi. (C) Larval arrest on control or F56A8.3 RNAi after N. parisii infection (dark gray) measured as the percent of animals reaching the L4 larval stage at 2 dpi. Uninfected animals grown on L4440 (control RNAi, light gray) are shown for comparison. Data are represented as mean values with SEM from seven independent experiments (****p<0.0001, unpaired two-tailed t-test). (D) Larval arrest on feeding RNAi after P. aeruginosa infection, quantified as the percent of animals reaching the L4 larval stage at 2 dpi. Data are representative of two independent experiments, with the mean and SEM from biological duplicates shown (n.s. is not significant and * is p<0.05 compared to L4440 in one-way analysis of variance with Dunnett’s multiple comparison test).