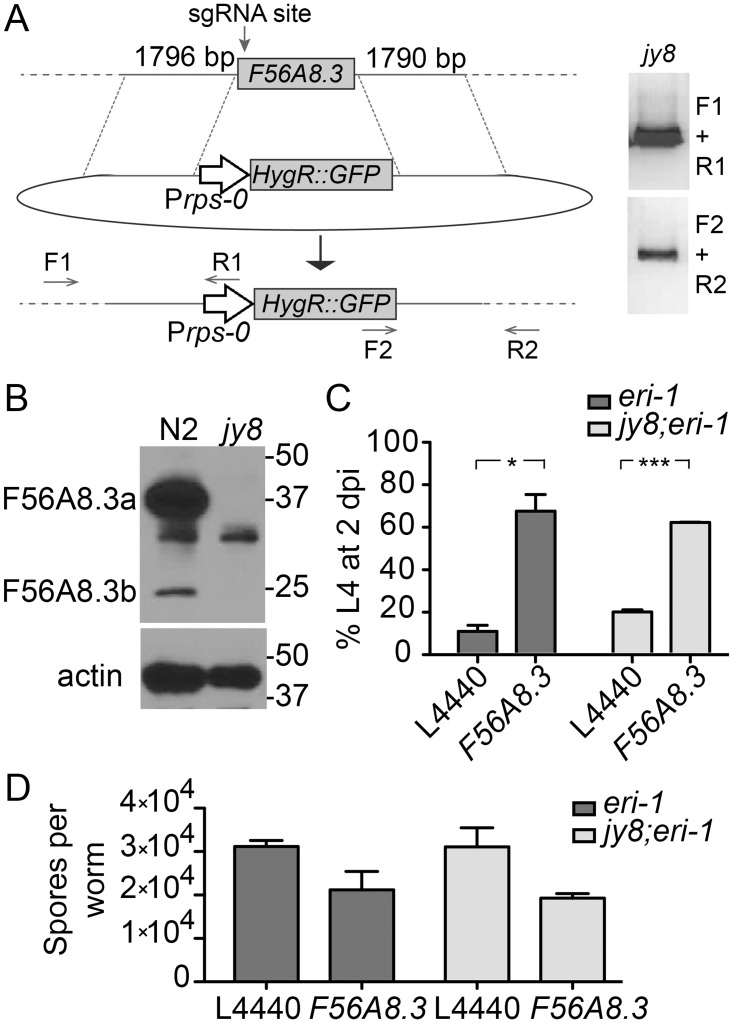
Fig 5. A complete deletion of the F56A8.3 locus does not recapitulate the larval arrest phenotype of F56A8.3 RNAi.
(A) Schematic representation of donor homologous repair template and predicted recombinant product after CRISPR-Cas9 mediated cutting of F56A8.3 (at the indicated sgRNA site). The donor template (middle) contains homologous regions of 1796 bp upstream and 1790 bp downstream of the F56A8.3 start and stop codons, respectively, which flank a Prps-0-controlled hygromycin-resistance gene (HygR) with an intergenic GFP expressed as part of an operon. Primer pairs F1+R1 and F2+R2 (bottom) flank the upstream and downstream genomic insertion sites, and their PCR products were detected in knockout mutant ERT425 F56A8.3(jy8) (right). (B) F56A8.3 protein in N2 and F56A8.3 knockout mutation ERT425 F56A8.3(jy8) detected with anti-F56A8.3. Indicated molecular weight markers are in kilodaltons (kD), and actin loading controls are shown (bottom). (C) Larval arrest of eri-1 and ERT430 F56A8.3(jy8) knockout (in an eri background) on control or F56A8.3 RNAi after N. parisii infection, measured as the percent animals reaching the L4 stage at 2 dpi. Data are represented as mean values with SEM from two independent experiments (*p = 0.021 (left), ***p = 0.0006 (right), unpaired two-tailed t-test). (D) Pathogen load at 40 hpi of eri-1 and ERT430 F56A8.3(jy8) knockout (in an eri-1 background) on control or F56A8.3 RNAi measured as the number of spores produced per animal. Data are represented as mean values with SEM from three biological replicates.