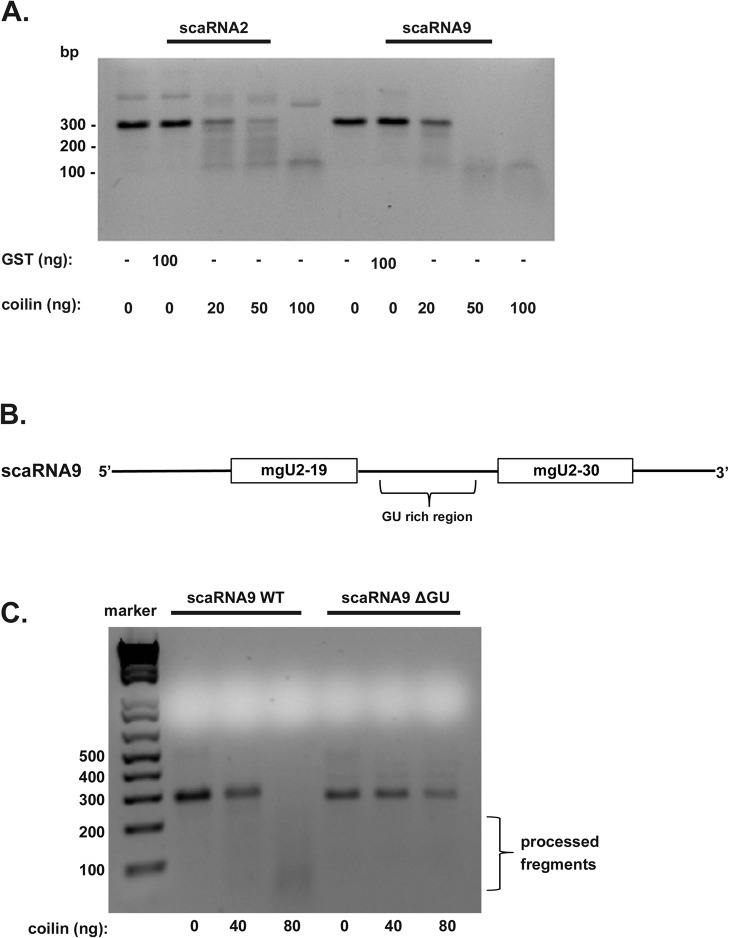
Fig 1. Coilin processing activity shows preference towards scaRNA9 compared to scaRNA2, and the GU region of scaRNA9 impacts coilin processing.
(A) RNA processing assay comparing the processing of in vitro transcribed scaRNA2 and scaRNA9 by bacterially purified coilin. Equivalent amounts of RNA were incubated with no protein, GST (100 ng) or increasing amounts of pure, nucleic acid free coilin (20 ng, 50 ng and 100 ng). Products were resolved on a 2% agarose gel and detected by ethidium bromide staining. (B) Schematic showing full-length scaRNA9 (353 nucleotides in length), the mgU2-19 and mgU2-30 RNAs that are derived from the full-length RNA, and the GU-rich region that lies in between these smaller guide RNAs. (C) RNA processing assay comparing the processing of in vitro transcribed scaRNA9 and scaRNA9 with a deleted GU-rich region (scaRNA9 ΔGU) by purified coilin. Approximately equal amounts of RNA were incubated with no protein, or two amounts of pure, nucleic acid free coilin (40 ng and 80 ng). Products were resolved on a 2% agarose gel and detected by ethidium bromide staining.