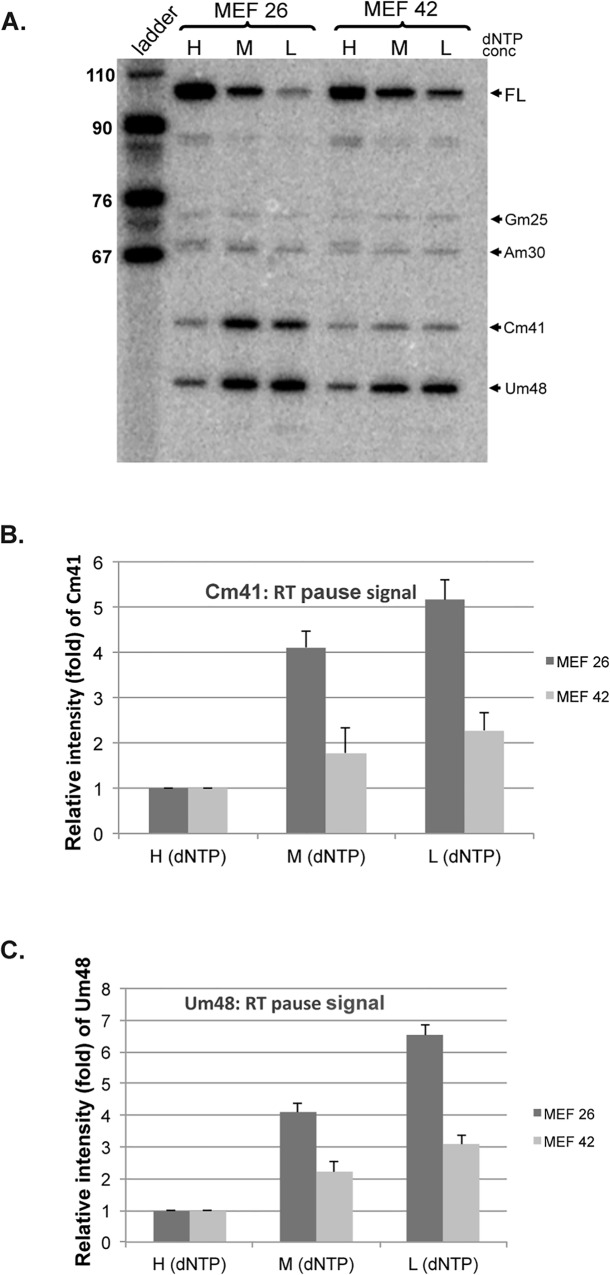
Fig 7. 2′-O-methylation of U2 snRNA is reduced in coilin knockout MEF cells.
(A) Primer extension experiments were done to map the 2’-O-methylation of individually modified nucleotides in U2 snRNA using equal amounts of RNA isolated from MEF 26 (coilin wild-type) and MEF 42 (coilin knock-out) cell lines. High, medium and low (H,M and L) deoxynucleoside triphosphate concentrations were used. The resulting pause signals corresponding to 2′-O-methylated residues in mouse U2 snRNA are indicated by each arrow. (B) Relative levels/intensities of Cm41 and Um48 in MEF26 and MEF 42 cell lines were calculated. For each lane, the percentage of the Cm41 and Um48 signals was calculated. The percentage values of Cm41 and Um48 of the medium and low dNTP lanes were then normalized to that of the high dNTP lanes. The ladder is shown in nucleotides.