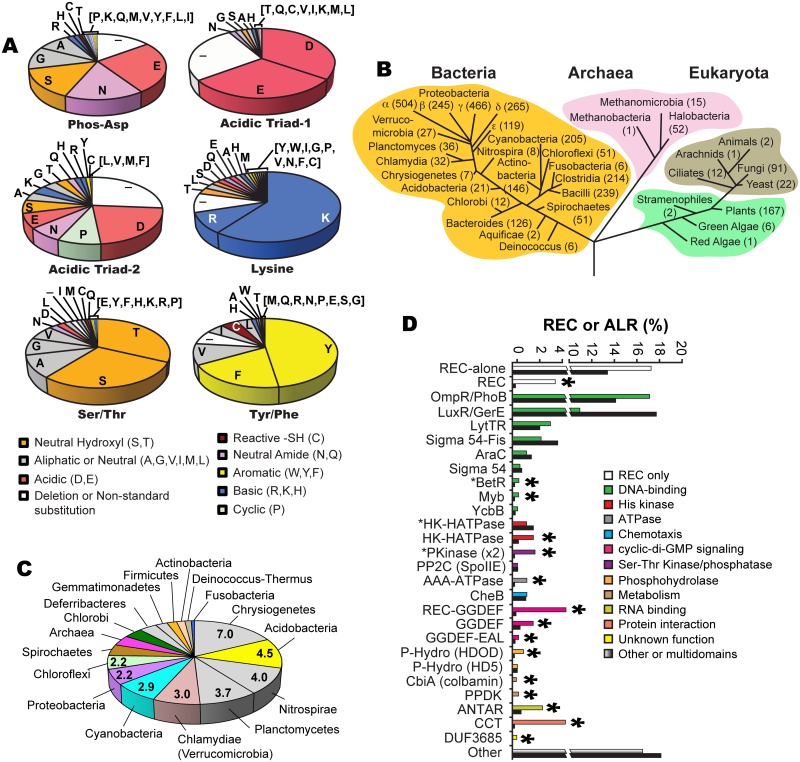
Fig 1. ALR statistics and phylogeny.
(a) Frequency of amino acid substitutions within six key ‘invariant’ REC residues in ALR sequences: the (now changed in ALRs) conserved histidine kinase phosphorylated aspartate residue position that defines the ALR subfamily (Phospho-Asp), acidic triad residue-1 (Glu9 in RitR) and acid triad residue-2 (Lys10 in RitR) that normally help coordinate the metal ion active pocket, the Tyrosine/Phenylalanine (Tyr/Phe, Tyr100 in RitR) and Threonine/Serine (Thr/Ser, Asp81 in RitR) that make up the Y/T-coupling system, and the conserved pocket Lys (Lys103 in RitR). Notice that where catalytic active pocket Asp/Lys residues have often been changed in ALR sequences (top panel), the T/Y-coupling residues generally remain conserved (bottom panel). This trend in conservation is also observed for the acidic triad-1 and the universally conserved pocket Lys residue (Lys103 in RitR), but not for acidic triad-2. (b) Taxonomic distribution of ALR sequences. The number of ALRs discovered in the given class or phylum is shown in parentheses. (c) Distribution of the average number of ALR sequences per completed genome by phyla. (d) Bar graph of the percentage contribution of a given Effector Domain (ED) within total REC sequences (shown as black bars) and ALR sequences only (shown as non-black bars). An asterisk above the bars indicates that ALRs are enriched for the ED by over 50% within the ALR population compared to their representation within typical REC sequence populations. An asterisk in front of the ED name indicates that the ALR or REC domain is (unusually) C-terminal to the ED sequence.