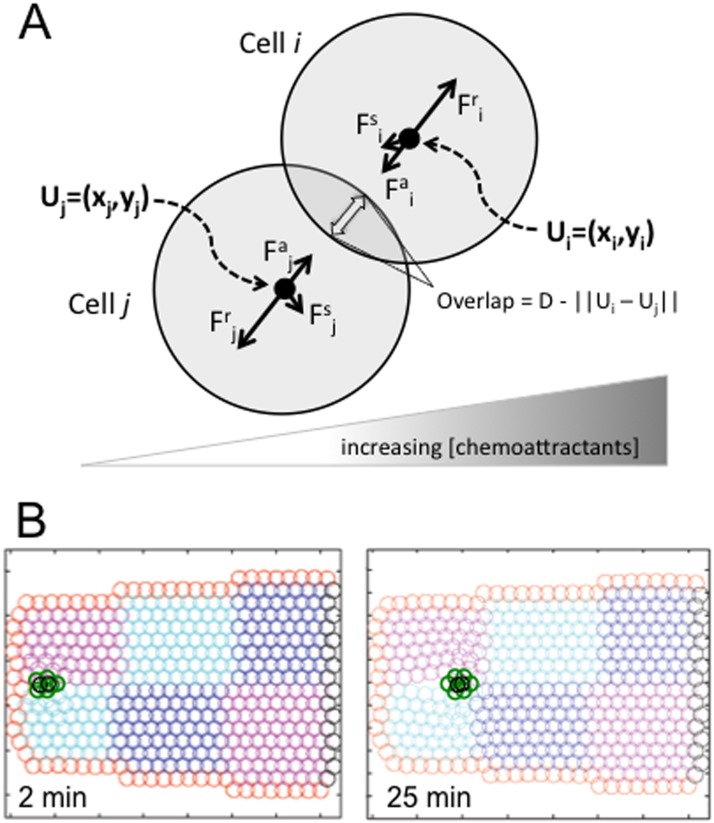
Fig 2. A force-based biophysical model comprised of discrete Individual Math Cells.
(a) The forces between two adjacent IMCs, i and j. The repulsive force acts to separate contacting IMCs, while the adhesive force acts to attract IMC i and j when they are within an ε distance from one another. If one of the IMCs is migratory, it produces a migratory force perpendicular to the axis of interaction due to signal from the gradient of chemoattractants. There is stochastic fluctuation in the position of each IMC. These forces balance and produce overlap between i and j of D − ‖U i − U j‖2, where D is the diameter of the IMC. Adhesion force between IMCs creates the integrity of a large single cell or cluster of individual cells with special affinity. The central cluster with a higher adhesion coefficient is closer or more tightly bound to one another than to outside cells, or than outside cells are to one another. (b) The IMC- based domain and simulation in two dimensions. The anterior half of the egg chamber is represented by IMCs with different properties. The epithelial cells are equated to IMCs (red), while the nurse cells are formed from many IMCs (blue, pink, and cyan). The line of black IMCs to the right is the surface of the oocyte at the mid-point of the whole egg chamber. The migratory cluster is formed of tightly bound border cells (green) and polar cells (black). 25 minutes into a 2D simulation, the model shows penetration of the cluster into the egg chamber between malleable nurse cells.