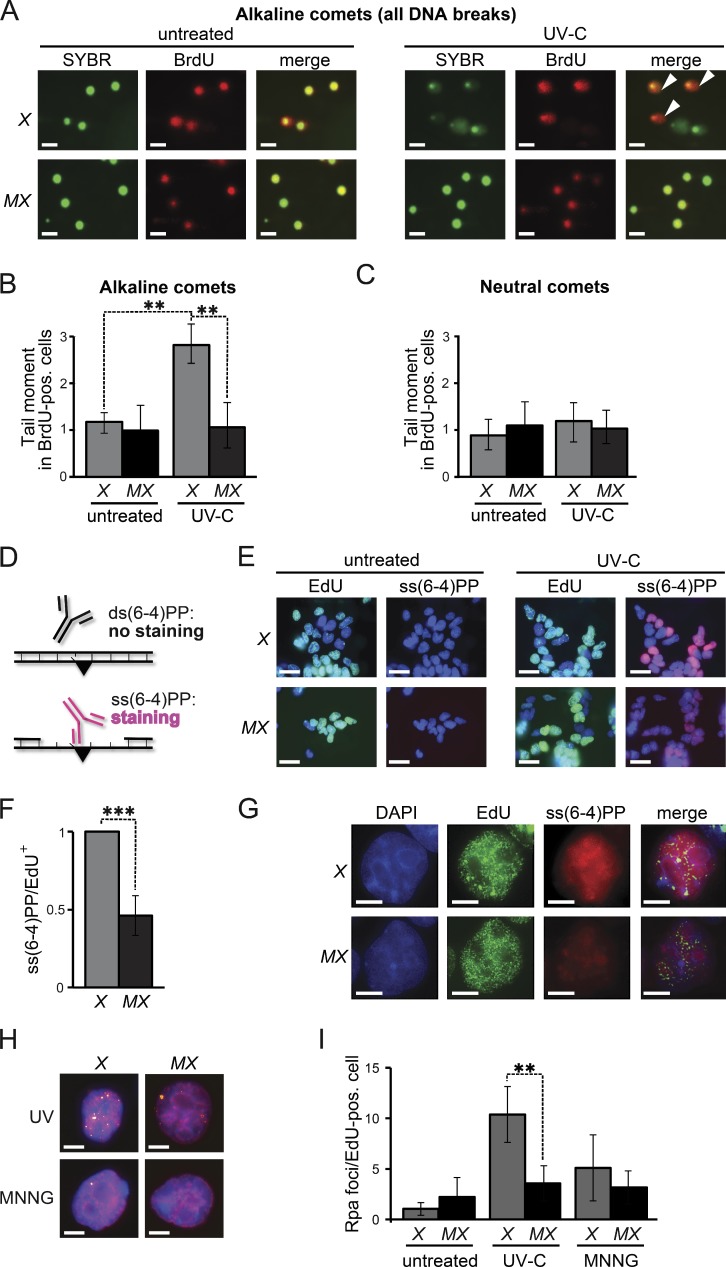
Figure 3.
Msh2/Msh6-dependent, S-phase-associated, induction of ss(6–4)PP. (A) Alkaline comet assays of nocodazole-treated (mitotic) cells 16 h after UVC-treatment (0.75 J/m2). Nuclei are stained with SYBR green. BrdU-positive nuclei were replicating during exposure. Comets (white arrowheads) are representative of ssDNA breaks. X, Xpa−/− ES cells, MX, Msh6−/−Xpa−/− line 4. Bars, 10 µM. (B) Quantification of tail moments, representing ssDNA breaks, in BrdU-positive mitotic cells after alkaline electrophoresis. Bars represent averages of three independent experiments. (C) Quantification of tail moments, representing dsDNA breaks, in BrdU-positive mitotic cells after neutral electrophoresis. Bars represent averages of three independent experiments. (D) Immunostaining to detect ss(6–4)PP (black triangle). (E) Staining for ss(6–4)PP (magenta) in ES cell lines, UVC treated (2 J/m2) during replication (EdU-positive nuclei, green), at 4 h after exposure. X, Xpa−/− ES cells; MX, Msh6−/−Xpa−/− line 4. Blue, nuclear stain (DAPI). Bars, 10 µM. (F) Relative levels of ss(6–4)PP in EdU-positive nuclei at 4 h after UVC exposure. Values were corrected for the 0 h time point. The ss(6–4)PP level in Xpa−/− ES cells was set to 1. Bars represent averages of three independent experiments. (G) Partial co-localization of Msh2/Msh6-dependent ss(6–4)PP with EdU (replication foci; pulse-labeled immediately after UVC exposure). Cells were stained at 4 h after UVC exposure. In the merged panels, the brightness of the EdU channel was reduced slightly. Bars, 2.5 µM. (H) Chromatin-associated Rpa foci (yellow) at 8 h after treatment of replicating (EdU positive, magenta) cells with UVC (0.75 J/m2), or with MNNG (4 µM) to induce canonical MMR. Blue, nuclear stain (DAPI). Bars, 2.5 µM. (I) Quantification of the numbers of Rpa foci in EdU-positive nuclei, at 8 h after UVC or MNNG treatment. One representative experiment of in total three experiments is shown.