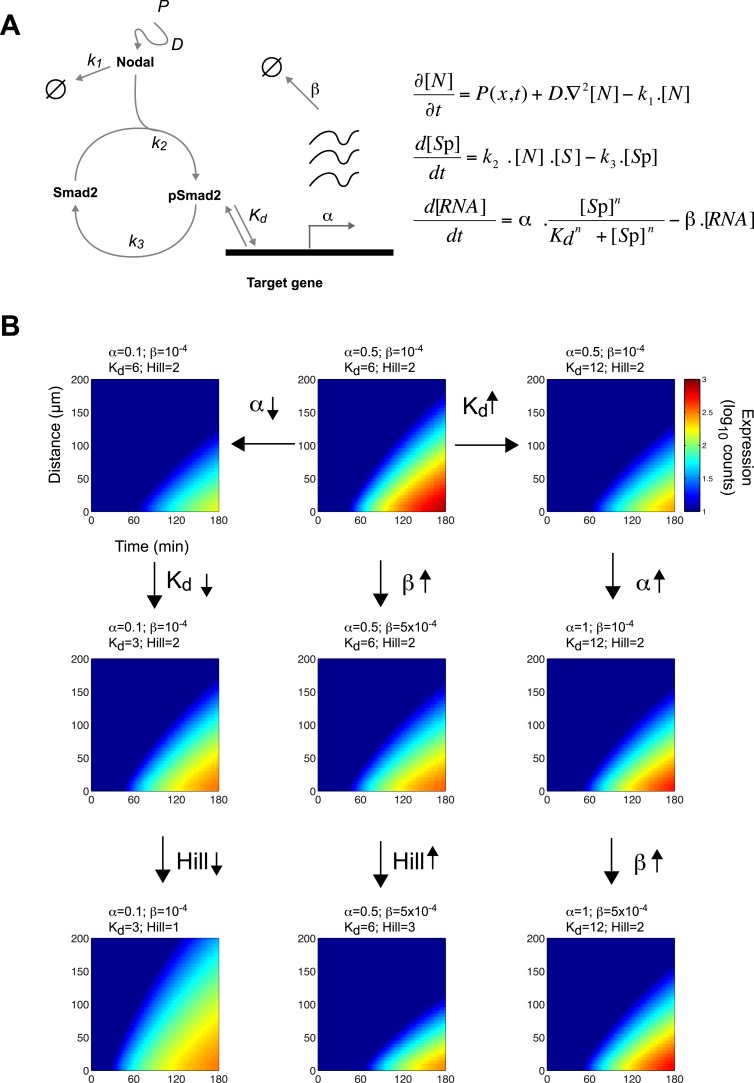
Figure 4. A kinetic model for Nodal morphogen interpretation.
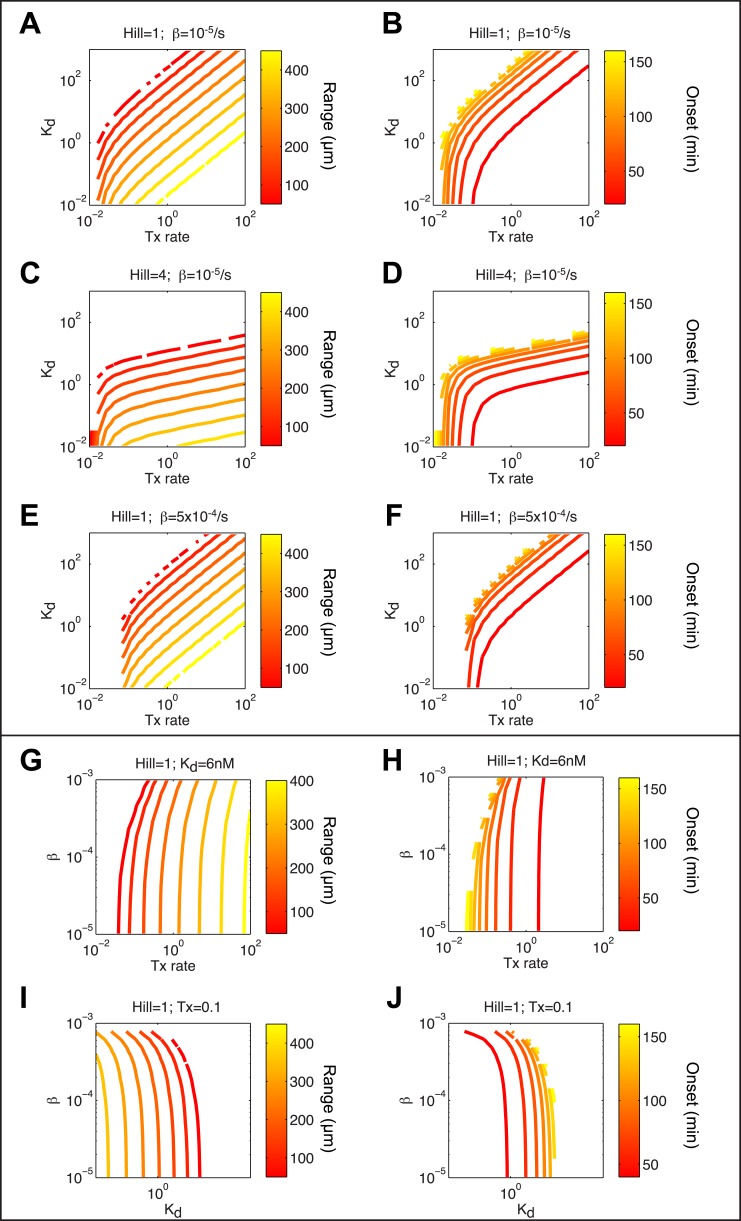
(A) Diagram of the Nodal signaling pathway used for modeling (left) and coupled differential equations describing the changes of Nodal, activated Smad2 and target genes over time (right). The Nodal ligand is locally produced, diffuses and via kinase receptors phosphorylates Smad2. Phosphorylated Smad2 acts as a transcription regulator and binds to target genes to induce transcription. (B) Spatiotemporal gene expression patterns were simulated over 3 hr using the kinetic model. Each panel depicts the expression pattern resulting from a unique combination of the four free parameters involved in mRNA production (transcription rate α, degradation rate β, dissociation constant Kd and Hill coefficient) while other parameters are held constant. Note how changes in these parameters change the range of target gene expression. See Figure 4—figure supplement 1 for more extensive simulations.