Figure 4.
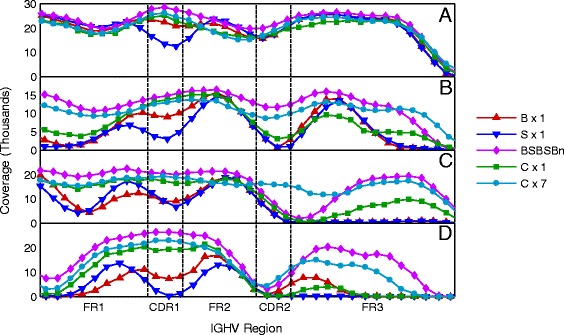
Iteration greatly increases IGHV read coverage from empirical SOLiD data. Coverage versus position within the FR1 to FR3 regions of the identified IGHV in four specimens with varying overall mutation rates. (A) Specimen 128, IGHV1-18, 7.3% mutation. (B) Specimen 134, IGHV3-48, 15.3% mutation. (C) Specimen 136, IGHV3-15, 17.3% mutation. (D) Specimen 132, IGHV1-46, 17.7% mutation. These plots show differences in read coverage between BFAST (B × 1), SHRiMP2 (S × 1), and CUSHAW3 (C × 1) mapping to germline sequences as the reference without iteration and the improvement in coverage obtained using iterative mapping wherein BFAST and SHRiMP2 are alternated twice, followed by additional BFAST iterations (BSBSBn) or CUSHAW3 for seven iterations (C × 7). Note how initial mapping coverage is lower in the CDR regions which typically are more highly mutated, but as the overall mutation rate increases, large regions including FR as well as CDR show severe loss of coverage due to the inability of the alignment programs to handle the clustered deviations from reference. See text for details.
