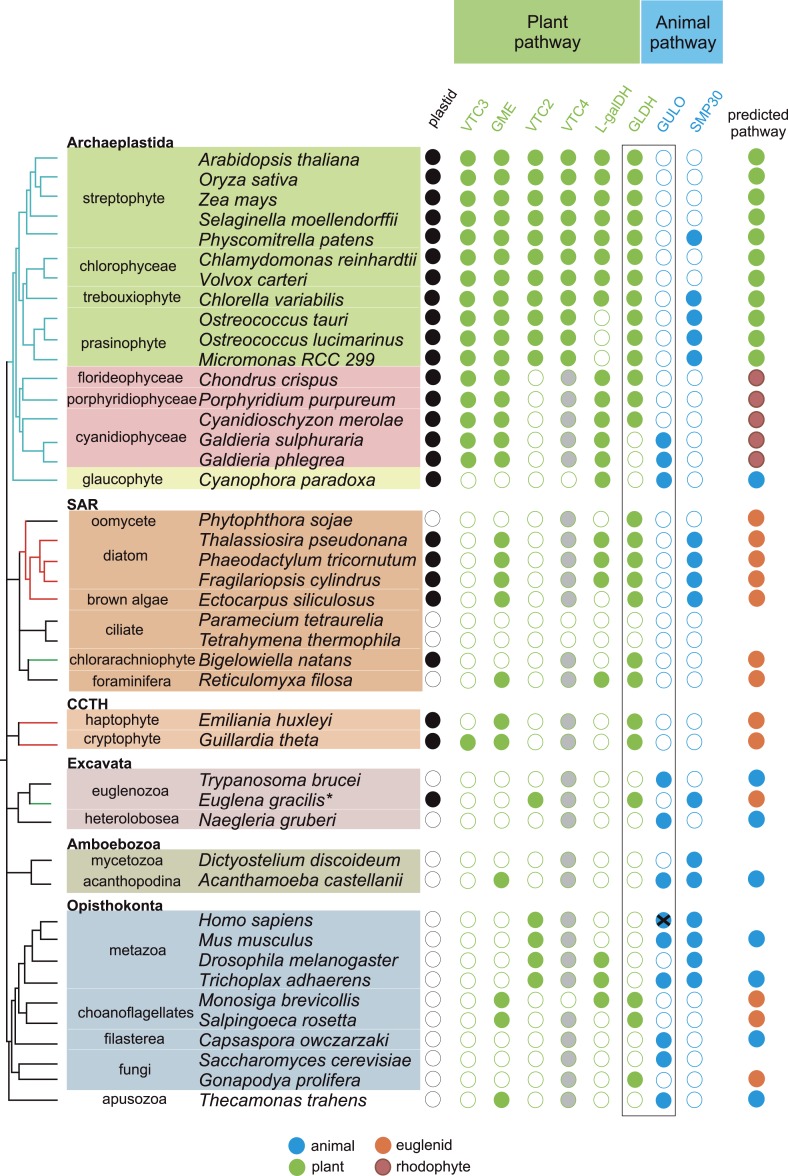
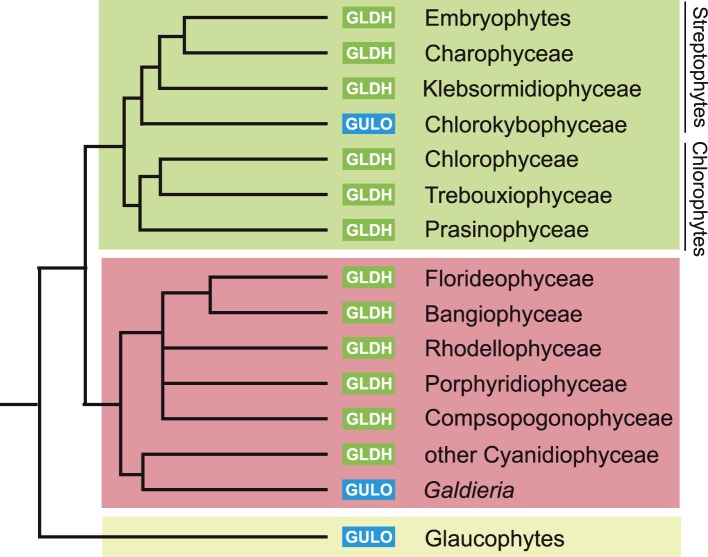
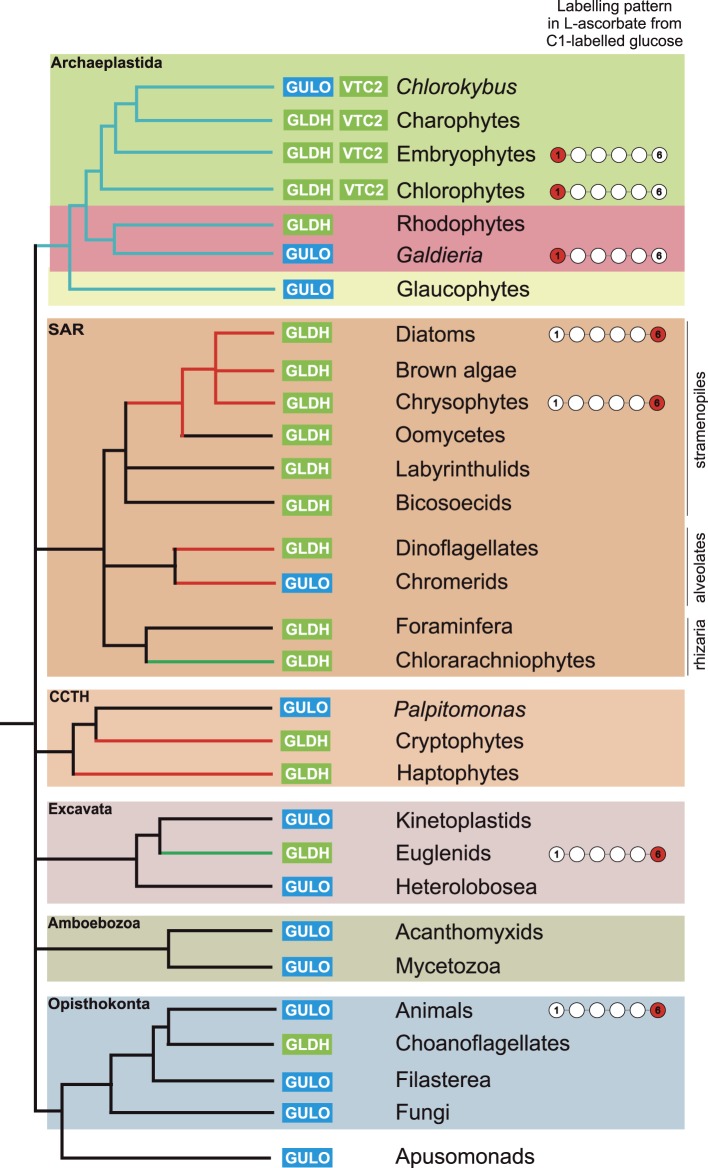
Figure 2. Coulson plot indicating the taxonomic distribution of the different ascorbate pathways.
40 eukaryote genomes were analysed for the presence of genes in the ascorbate biosynthetic pathways. The two potential terminal enzymes in the pathway are boxed. GLDH is common to both the ‘plant’ and ‘euglenid’ type pathways. A schematic tree depicts the currently accepted phylogenetic relationships between organisms. The predicted route of ascorbate biosynthesis in each organism is shown. Note that ‘euglenid’ and ‘rhodophyte’ type pathways cannot currently be distinguished from sequence analysis alone and the predictions are based on biochemical evidence. Asterisk denotes a genome assembly was not available for Euglena gracilis and its transcriptome was analysed (‘Materials and methods’). Grey circles in VTC4 represent the presence of a highly similar enzyme, myo-inositol-1-phosphate phosphatase that exhibits l-galactose-1-phosphatase activity. GULO in trypanosomes and yeasts acts to oxidise the alternative substrates l-galactonolactone or D-arabinonolactone respectively. VTC3 is not a biosynthetic enzyme, but represents a dual function Ser/Thr protein kinase/protein phosphatase 2C that may play a regulatory role in the plant pathway (Conklin et al., 2013). Black cross represents a pseudogene encoding a non-functional enzyme. Organisms with sequenced genomes that were found to lack both of the terminal enzymes in the known pathways (GULO and GLDH) are likely to be ascorbate auxotrophs and were not included in the plot. These include Giardia intestinalis, Trichomonas vaginalis, Entamoeba invadens, Plasmodium falciparum and Perkinsus marinus.