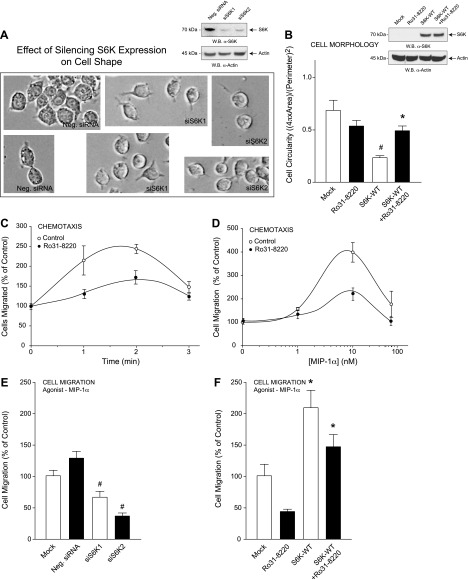
Figure 4.
S6K-mediated cell shape changes contribute to cell migration. A, B) RAW264.7 macrophages were transfected with 300 nM siRNAs specific for either S6K1 or S6K2 (A) or 2 µg S6K-WT plasmid for 3 days using electroporation and then mock-treated or treated with 10 nM of the S6K-specific inhibitor Ro31-8220 for 20 minutes (B); subsequent cell lysates were used for WB (insets, respectively). A) Bright-field photomicrographs of RAW264.7 macrophages were imaged at 72 hours post-transfection and are presented as duplicate fields that indicate cells became more circular in appearance following silencing of S6K. C, D) Optimization of MIP-1α–mediated chemotaxis as a function of time (C) and MIP-1α concentration (D) in the absence or presence of 10 nM of the S6K-specific inhibitor Ro31-8220. Results are presented as the mean number of cell migrated compared with the control ± sem of triplicate samples. E, F) Samples in (A) and (B) were also used for MIP-1α–mediated chemotaxis in response to 10 nM MIP-1α for 1 h. *P < 0.05, statistically significant increase, between samples and controls. #P < 0.05, statistically significant decrease, between samples and controls.