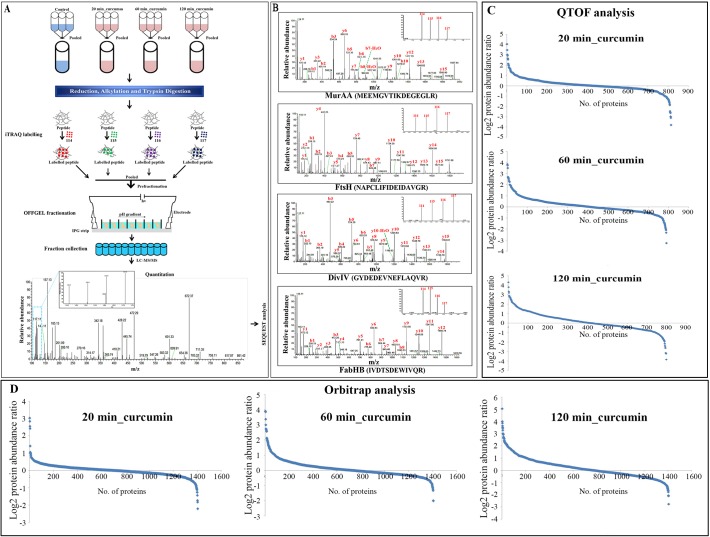
Fig 3. Schematic representation of experimental strategy for temporal proteome analysis of B. subtilis under curcumin treatment by iTRAQ-based quantitative proteomics.
(A) Samples processed in triplicate were pooled from control, 20, 60 and 120 min curcumin treated cultures and labelled with iTRAQ reagent 114, 115, 116 and 117, respectively. The labelled peptides were fractionated in OFFGEL fractionators using high resolution (24 cm; 3–10 pH) IPG strips and each fraction was desalted using C18 tips. Desalted fractions were subjected to LTQ-Orbitrap Velos mass spectrometer for protein identification and quantitation. (B) Representative MS/MS spectrum of a few selected differentially expressed proteins identified after curcumin treatment. UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurAA), ATP-dependent zinc metalloprotease FtsH, Septum site-determining protein (DivIVA), and 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1 (FabHB). Inset showing the iTRAQ reporter ion intensities for representative peptides in control and curcumin treated samples. (C) S-curve analysis exhibiting distribution of the differentially expressed proteins in B. subtilis after 20, 60 and 120 min of curcumin treatment identified using Q-TOF (average of three triplicate runs). (D) S-curve analysis exhibiting distribution of the differentially expressed proteins in 20, 60 and 120 min curcumin treated B. subtilis identified using LTQ-orbitrap.