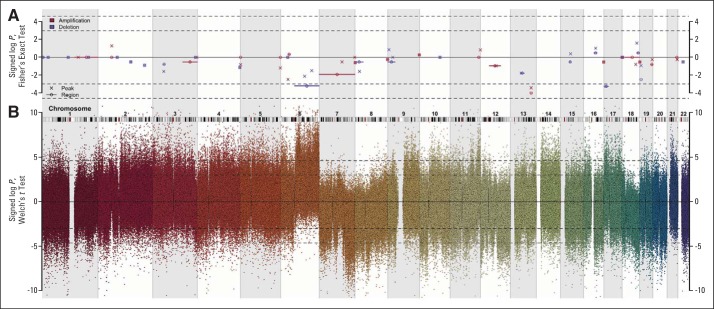
Fig A4.
Characterization of the genotypes of B-cell–associated gene signature subtypes. Copy number aberrations (CNAs) in the assigned subtypes were found at various chromosome positions when studied in 165 samples from Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project (MDFCI) with Affymetrix Genome-Wide Human SNP Array 6.0 and Affymetrix Gene Chip Human Genome U133 Plus 2.0 Arrays or U133A+B Arrays. (A) Signed log P values calculated using Fisher's exact test for the hypothesis of equal proportions of CNA in germinal center B-cell–like (GCB) centrocyte (CC; n = 32) versus GCB-centroblast (CB; n = 23) in peaks and regions (as defined by Monti et al13) are plotted against the chromosomal position of the region. Crosses and circles denote genomic peaks and regions, respectively. Color indicates the type of CNA investigated, as follows: amplification (red) or deletion (blue). Values greater than zero indicate more frequent CNAs in GCB-CC compared with GCB-CB, and vice versa for negative values. The dashed horizontal lines indicate the 95% and 99% acceptance intervals. (B) Signed log P values calculated based on t test for the hypothesis of no difference in copy number for each probe set in GCB-CC and GCB-CB samples are plotted against the chromosomal position of that probe set. Values greater than zero indicate amplification in GCB-CC or deletion in GCB-CB, and vice versa for values less than zero.