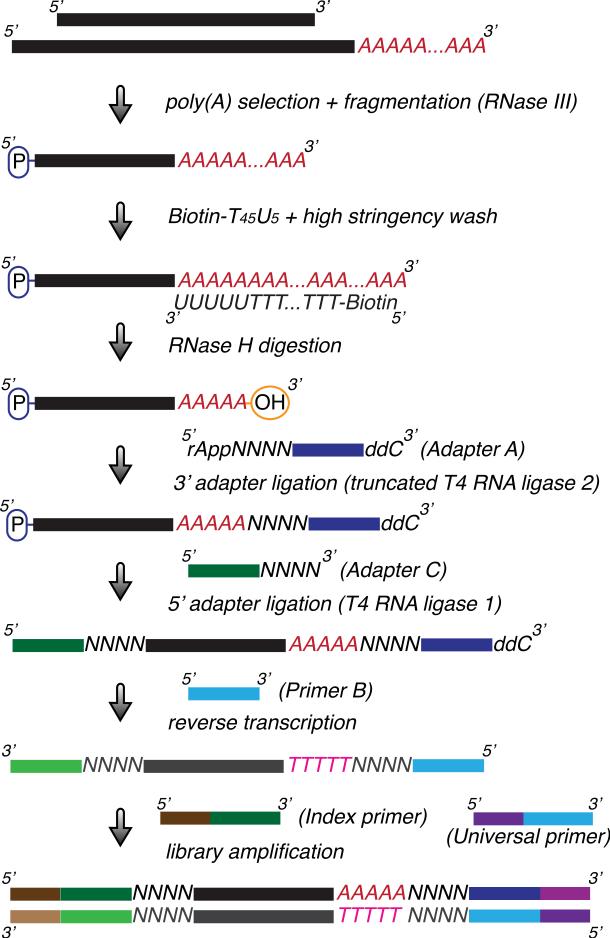
Figure 1.
Schematic of an indexed DNA library preparation for sequencing poly(A) sites (3’ mRNA isoforms) on a genomic scale. Poly(A)-tailed RNAs are first enriched and fragmented by RNase III digestion. 3’ polyadenylated RNA fragments captured by Dynabeads coated with a Biotin-T45U5 hybrid DNA/RNA oligo are subjected to RNase H digestion to generate RNA fragments containing 3’ termini with only a few remaining As. After ligation of linkers (Adapters A and C) to both ends, the resulting RNA fragments are reverse-transcribed to yield single-stranded cDNA fragments using a primer (Primer B) based on the sequence of Adapter A. The cDNA fragments are in turn amplified with a universal primer (Universal primer) and a second primer containing the desired index (e.g., Index primer 1) to generate a double-stranded DNA library. The quality and quantity of the purified DNA library can be examined using the Agilent Bioanalyzer and Quantitative-PCR.