Figure 2.
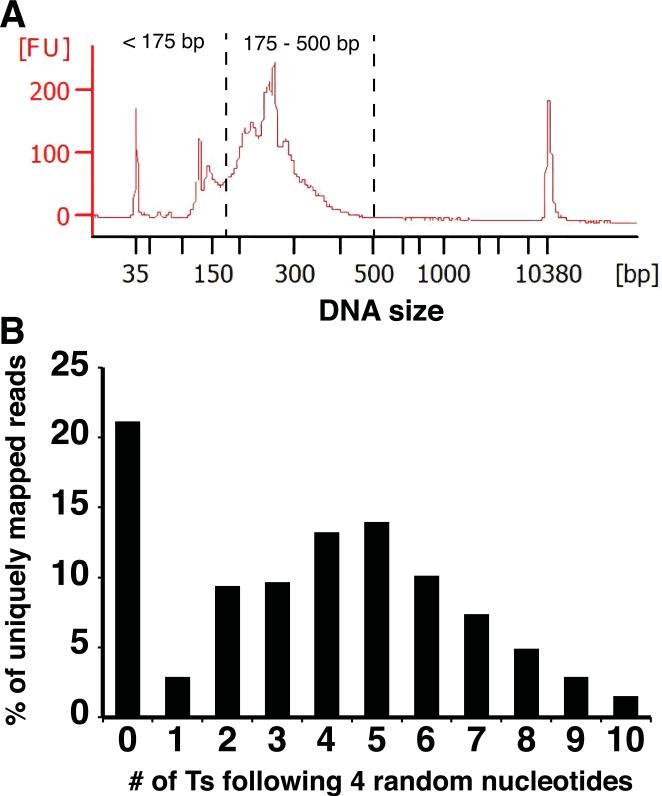
(A) Agilent Bioanalyzer analysis (High-sensitivity DNA Kit) of a SPRI-beads-purified DNA library is shown. DNA fragments smaller than 175 bp include library DNAs with very short cDNA inserts, PCR primer dimers, and residual PCR primers. Gel purification is required to further extract only DNA fragments between 175 bp and 500 bp for Illumina sequencing. The DNA peaks at 35 bp and 10380 bp are high sensitivity DNA markers used in Agilent Bioanalyzer analysis. (B) Distribution of the number of consecutive Ts following four random nucleotides in uniquely mapped sequence reads. The stretch of Ts corresponds to the As remaining from poly(A) tails after RNase H digestion. Reads with no Ts are likely to be caused by RNase H over-digestion and contaminating non-3’ RNA fragments.