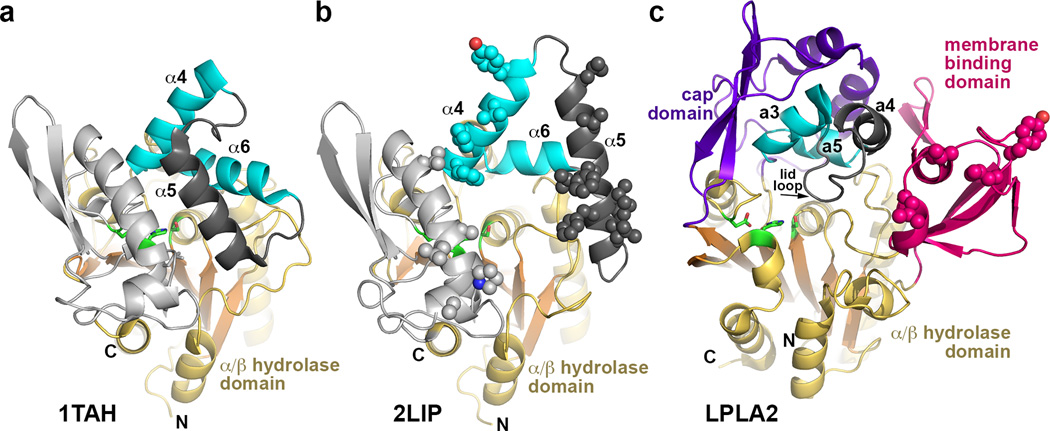
Figure 2.
Structural comparison of Family I triacylglycerol lipases with LPLA2. These enzymes all feature a similar α/β hydrolase core and cap domains that contain a topologically and structurally similar motif consisting of two helices (cyan) joined by a flexible loop (dark grey). In the bacterial lipase family, the α5 helix within this loop functions as an active site lid in the closed state and as a membrane-binding element in the open state. (a) Closed conformation of triacylglycerol lipase from Pseudomonas glumae (PDB entry 1TAH). (b) Open conformation of Pseudomonas cepacia lipase (PDB entry 2LIP). Potential membrane binding residues are shown as spheres. (c) In comparison, LPLA2 seems to exhibit an open conformation, and the membrane binding domain occupies a similar topological location with respect to the active site as the α5 helix in its open configuration in panel B. Hydrophobic residues shown to be involved in membrane binding are shown as spheres. The lid loop and subsequent a4 helix of the cap domain (dark gray) is topologically equivalent to α5 in panels A and B.