Abstract
MicroRNAs (miRNAs) are short, endogenous RNA molecules that have essential roles in regulating gene expression. They control numerous physiological and cellular processes, including normal bone organogenesis and homeostasis, by enhancing or inhibiting bone marrow cell growth, differentiation, functional activity and crosstalk of the multiple cell types within the bone. Hence, elucidating miRNA targets in bone marrow stromal cells has revealed novel regulations during bone development and maintenance. Moreover, recent studies have detailed the capacity for bone stromal miRNAs to influence bone metastasis from a number of primary carcinomas by interfering with bone homeostasis or by directly influencing metastatic tumor cells. Owing to the current lack of good diagnostic biomarkers of bone metastases, such changes in bone stromal miRNA expression in the presence of metastatic lesions may become useful biomarkers, and may even serve as therapeutic targets. In particular, cell-free and exosomal miRNAs shed from bone stromal cells into circulation may be developed into novel biomarkers that can be routinely measured in easily accessible samples. Taken together, these findings reveal the significant role of bone marrow stroma-derived miRNAs in the regulation of bone homeostasis and bone metastasis.
Biogenesis and function of miRNAs
MicroRNAs (miRNAs) are a large family of noncoding ∼22-nucleotide-long RNA molecules that negatively regulate gene expression.1 It is estimated that miRNAs regulate about 50% of all protein-coding genes.2 They repress gene expression through complementary binding to sites in the 3′-untranslated region (UTR) of target mRNAs.3,4 miRNA-mediated inhibition occurs either by mRNA degradation or translational silencing. Cleavage of target mRNAs occurs when the miRNA and the target mRNA exhibit perfect complementarity.3 Conversely, miRNA–mRNA pairings with imperfect complementarity result in translational inhibition in the absence of target cleavage.3,5 Thus, even in the absence of perfect binding, the association between an miRNA and a target 3′-UTR still results in the suppression of a target protein. Owing to the relatively short length of the seed sequence that binds to the 3′-UTR (5–7 nucleotides), each miRNA can potentially recognize hundreds of mRNAs and is thus a powerful molecular manager that can control several gene regulatory networks simultaneously.
miRNA genes are typically transcribed into stem-loop structures from intra- or intergenic regions by RNA polymerase II and undergo sequential cleavage steps to produce mature miRNAs.2 In the nucleus, newly transcribed pri-miRNAs (primary miRNAs) are cleaved by a complex formed by the RNase III enzyme Drosha and the double-stranded RNA-binding protein Pasha (or DGCR8) to produce precursor miRNAs. These precursor miRNAs are about 70–100 nucleotides long.6 They are exported from the nucleus by Exportin 5 and the Ran-GTP cofactor,7 where they undergo a second cleavage by the endoribonuclease Dicer to produce a double-stranded, ∼18–25-nucleotide-long mature miRNA. One miRNA strand then combines with Argonaute (AGO2) proteins to form the RNA-induced silencing complex, allowing for directed pairing with target mRNAs and mediating rapid fine-tuning of gene expression.4
miRNA regulation within bone stromal cells
Bone formation and turnover are complex processes that involve differentiation and crosstalk of multiple cell types for the generation and remodeling of the skeleton.8,9,10 Inhibition of mRNA translation by miRNAs has emerged as an important regulator of developmental osteogenic signaling pathways, osteoblast growth and differentiation, osteoclast-mediated bone resorption activity and general bone homeostasis. Hence, it is not surprising that cell-type-specific disruption of miRNA biogenesis by deletion of Dicer would have a significant impact on physiological bone formation and remodeling (Table 1). For example, in vivo ablation of Dicer in osteoprogenitors, osteoblasts and chondrocytes by Col1a1-Cre resulted in severe skeletal deformities in fetal mice as it inhibited their maturation into cells that lay down bone mineral.11 The mice had a disproportione cartilage skeleton and a reduction in mineralized tissue at E14.5. Excision of Dicer in mature osteoblasts by Osteocalcin-Cre produced a viable mice, which exhibited delayed bone development that corresponded with reduced osteoblast numbers.11 Similarly, Dicer deletion in chondrocytes, using Col2a1-Cre mice, modulated the proliferation and differentiation of cells into postmitotic hypertrophic chondrocytes leading to significant skeletal defects.12 Osteoclast-specific Dicer knockout, using CD11b-Cre or CTSK-Cre transgenic mice, disrupted osteoclastogenesis and reduced the number and activity of multinuclear osteoclasts leading to an increase in bone mass.13,14,15 Cultured bone marrow from these transgenic mice was also incapable of producing osteoclasts ex vivo, further confirming the defect in osteoclast differentiation.14,15 Importantly, this defect was not confined to Dicer; ablation of DGCR8 and AGO2 using small interfering RNAs (siRNAs) similarly resulted in decreased osteoclast maturation and bone resorption, and osteoclast-specific DGCR8 knockout mice display impaired bone development (Table 1).13,16 Taken together, these results reveal the necessity for proper miRNA regulation during the differentiation and maintenance of multiple essential bone marrow cell types required for bone formation and homeostasis.
Table 1. Key miRNA regulators of bone organogenesis and homeostasis.
| MiRNA biogenesis | Regulatory function | Activity | Strategy for reduced expression | Reference |
|---|---|---|---|---|
| Ago2 | Essential for development | Knockout inhibits osteoclast development | siRNA (in vitro) | 13 |
| DGCR8 | Essential for development | Knockout inhibits osteoclast development | CTSK-Cre, siRNA (in vitro) | 13,16 |
| Dicer | Essential for development | Knockout inhibits osteoblast, osteoclast, chondrocyte development | Col1a1-Cre, osteocalcin-Cre, CD11b-Cre, CTSK-Cre, Col2a1-Cre | 11,15 |
| Regulatory function | Activity | Targets | Reference | |
| Osteoclast | ||||
| MiR-21 | Activates | Promotes differentiation | PDCD4 | 14 |
| MiR-29b | Activates and/or inhibits | Activates and/or inhibits differentiation | Cdc42, Srgap2, Fos, Mmp2 | 26,27 |
| MiR-31 | Essential for development | Essential for differentiation | Rhoa | 28 |
| MiR-34a | Activates | Promotes differentiation | Tgif2 | 33 |
| MiR-125a | Inhibits | Inhibits differentiation | TRAF6 | 30 |
| MiR-146a | Inhibits | Inhibits differentiation, protects joint destruction during collagen-induced arthritis | 64 | |
| MiR-148a | Activates | Promotes differentiation | MAFB | 65 |
| MiR-155 | Inhibits | Inhibits differentiation | SCOS1, MITF | 31,32 |
| MiR-223 | Activates and/or inhibits | Positive and negative regulator of differentiation | 13,25 | |
| MiR-503 | Inhibits | Inhibits differentiation | RANK | 29 |
| Osteoblast | ||||
| MiR-15b | Activates | Promotes differentiation | Smurf1 | 66 |
| MiR-17∼92 | Activates | Promotes proliferation and differentiation | 67 | |
| MiR-17-5p, miR-106 | Inhibits | Inhibits differentiation | BMP2 | 68 |
| MiR-20a | Activates | Promotes differentiation | PPARγ, Bambi, Crim1 | 69 |
| MiR-23a/27a/24-2 | Inhibits | Inhibits apoptosis | FAK, Runx2, Satb2 | 21,22 |
| MiR-29a | Activates | Promotes differentiation | Osteonectin, Dkk1, Kremen2, sFRP2 | 70,71,72 |
| MiR-30c | Inhibits | Inhibits differentiation | Smad1, Runx2 | 21 |
| MiR-34c | Inhibits | Inhibits proliferation and differentiation | SATB2, Runx2, Notch pathway | 21,34,35 |
| MiR-93 | Inhibits | Inhibits mineralization | Sp7/Osx | 73 |
| MiR-100 | Inhibits | Inhibits differentiation | BMPR2 | 74 |
| MiR-125b | Inhibits | Inhibits differentiation | ErbB2, Sp7/Osx | 75,76 |
| MiR-133a | Inhibits | Inhibits differentiation | Runx2 | 21 |
| MiR-135a | Inhibits | Inhibits differentiation | Runx2 | 21 |
| MiR-137 | Inhibits | Inhibits differentiation | Runx2 | 21 |
| MiR-138 | Inhibits | Inhibits differentiation | FAK, indirectly Sp7/Osx | 24 |
| MiR-141, miR-200a | Inhibits | Inhibits differentiation | Dlx5 | 77 |
| MiR-143 | Inhibits | Inhibits differentiation | Sp7/Osx | 78 |
| MiR-155 | Inhibits | Regulates TNFα inhibition of osteoblast differentiation | SOCS1, MITF | 31 |
| MiR-181a | Activates | Promotes differentiation | Tgfbr1, Tgfbi | 79 |
| MiR-182 | Inhibits | Inhibits proliferation and differentiation | Foxo1 | 80 |
| MiR-204/211 | Inhibits | Inhibits differentiation | Runx2, Sost | 21 |
| MiR-205 | Inhibits | Inhibits differentiation | Runx2 | 21 |
| MiR-206 | Inhibits | Inhibits differentiation | Cx43 | 23 |
| MiR-208 | Inhibits | Inhibits differentiation | Ets1 | 81 |
| MiR-214 | Inhibits | Inhibits activity and bone formation | ATF4, Sp7/Osx | 82,83 |
| MiR-217 | Inhibits | Inhibits differentiation | Runx2 | 21 |
| MiR-218 | Inhibits | Inhibits differentiation | Runx2, Dkk2, Sfrp2 | 21 |
| MiR-322 | Activates | Promotes differentiation | Tob2, indirectly Sp7/Osx | 84 |
| MiR-335-5p | Activates | Promotes differentiation | DKK1 | 85 |
| MiR-338 | Inhibits | Inhibits differentiation | Runx2 | 21 |
| MiR-542-3p | Inhibits | Inhibits differentiation | BMP7 | 86 |
| MiR-637 | Inhibits | Inhibits differentiation | Sp7/Osx, Col4a1, Bmp2k | 87,88 |
| MiR-764-5p | Inhibits | Inhibits differentiation | CHIP/STUB1 | 89 |
| Regulatory function | Activity | Targets | Reference | |
| Chondrocytes | ||||
| Let-7 | Activates | Promotes proliferation | Cdc34, E2f5 | 90 |
| MiR-17–92 | Essential for development | Loss induces microcephaly and other skeletal defects in patients | 91 | |
| MiR-34a | Inhibits | Inhibits differentiation | Apoptosis | 92,93 |
| MiR-140 | Essential for activity | Essential for proper activity | Sp1, HDAC4, Pdgfra, Dnpep,Smad3, Rala | 17,18 |
| MiR-145 | Inhibits | Inhibits differentiation | Sox9 | 20 |
| MiR-199a* | Inhibits | Inhibits differentiation | Smad1 | 19 |
| MiR-221 | Inhibits | Inhibits proliferation | Mdm2 | 94 |
| MiR-335 | Activates | Promotes differentiation | Rock1, Daam1 | 95 |
| MiR-337 | Inhibits | Inhibits differentiation | Tgfbr2 | 96 |
| MiR-449 | Inhibits | Inhibits differentiation | Lef1 | 97 |
| MiR-1247 | Inhibits | Inhibits differentiation | Sox9 | 98 |
Abbreviations: Ago2, argonaute; DGCR8, double-stranded RNA-binding protein Pasha; miR, microRNA; siRNA, small interfering RNA; TNFα, tumor necrosis factor-α.
miRNA regulation in bone homeostasis
Similar to studies investigating the effects of disruption of the general miRNA biogenesis pathway, several recent reports have revealed the capacity for individual miRNAs to direct the differentiation and activity of cells residing in the bone microenvironment. In this context, miRNAs can serve as both negative and positive regulators during differentiation and functional activation by disrupting the temporally organized coordination of transcription factors and regulatory proteins (Table 1). Characterization of miRNAs that operate through tissue-specific transcription factors as well as intricate feed-forward and reverse loops has provided novel insights into the supervision of signaling pathways and regulatory networks controlling normal bone development and turnover.
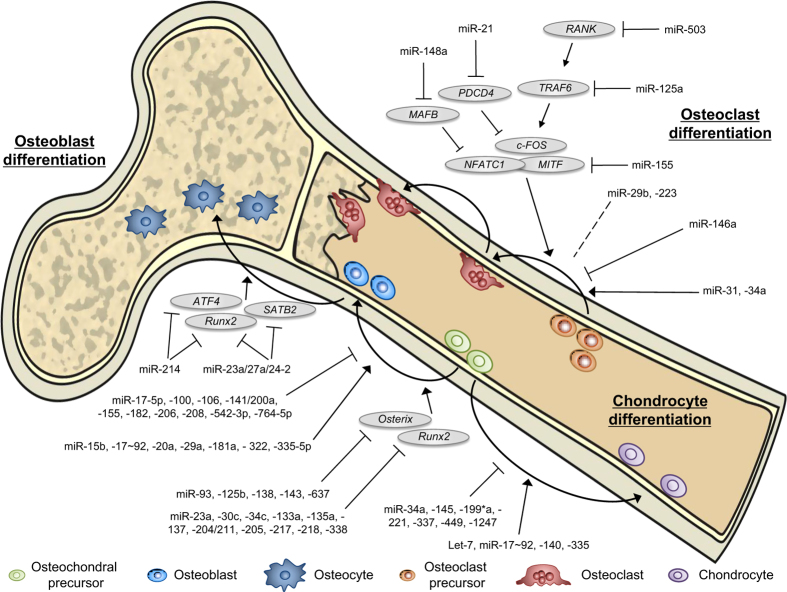
Chondrocytes are intricately regulated by a series of miRNAs (Figure 1 and Table 1), including miR-140, miR-199a* and miR-145. MiR-140 positively regulates chondrocyte differentiation by inhibiting HDAC4 and Dnpep, and contributes to craniofacial development and endochondral bone formation.17,18 Consistent with this, mice lacking miR-140 display accelerated bone formation at the embryonic and neonatal stages of development.18 Conversely, miR-199a* and miR-145 negatively regulate chondrogenesis; miR-199a* downregulates the expression of Smad1,19 whereas miR-145 targets Sox9, which encodes an essential transcription factor for chondrocyte differentiation.20
Figure 1.
MicroRNA (miRNA) involvement in bone homeostasis. MiRNA-mediated regulation of osteoclast, osteoblast, and chondrocyte differentiation and activity is essential for proper bone development and homeostasis. Known positive and negative miRNA regulators and several of their key targets are indicated.
miRNA-based regulation was observed during osteoblast differentiation and activation (Figure 1 and Table 1). Runx2, a member of the Runt transcription factor family, is essential for osteogenesis and is subject to regulation by a number of miRNAs, including miR-23a, miR-30c, miR-34c, miR-133a and miR-204.21 The miR-23a/-27a/24-2 cluster is tied to osteogenesis through an intricate Runx2 regulatory loop. Runx2 transcriptionally represses the expression of the miR-23a/-27a/24-2 cluster by binding to its promoter region. On the other hand, miR-23a is capable of directly repressing Runx2 expression.22 Thus, Runx2-mediated inhibition of the miR-23a/-27a/24-2 cluster initiates a positive feedback loop. Moreover, inhibition of this miRNA cluster also derepresses another activator, SATB2, allowing for progression of the cell through osteogenesis.22 MiR-206 was also shown to inhibit osteoblast development by targeting Cx43, whereas miR-206 knockdown promoted differentiation.23 MiR-138 inhibited osteogenesis by directly targeting signaling through focal adhesion kinase leading to attenuated bone formation in vivo.24 Importantly, treatment of human mesenchymal stem cells (MSCs) with anti-miR-138 oligonucleotides increased osteogenic capacity and bone formation.24
Several miRNAs were shown to exhibit dichotomous roles during osteoclastogenesis by simultaneously enhancing and suppressing differentiation (Figure 1 and Table 1), including miR-223 (Sugatani and Hruska13,25) and the miR-29 family.26,27 Although one study reported that ectopic miR-29b expression inhibited osteoclast activity,26 a more recent report found miR-29 to be able to promote osteoclastogenesis by directly targeting Cdc42 and Srgap2, and knockdown of miR-29 in preosteoclasts to inhibit differentiation.27 Additional studies uncovered miRNAs with more easily defined roles during osteoclast differentiation (Figure 1 and Table 1). MiR-31, which is highly upregulated during osteoclast development, has been shown to be essential for proper differentiation,28 whereas miR-21 positively regulated osteoclastogenesis by targeting the c-Fos inhibitor PDCD4.14 Conversely, RANK targeting by miR-503 inhibited osteoclastogenesis, and silencing miR-503 enhanced in vivo bone resorption.29 MiR-125a was shown to inhibit differentiation by suppressing TRAF6 expression,30 and miR-155 regulated cell-fate commitment within macrophages through the downregulation of SOCS1 and MITF, thereby effectively inhibiting differentiation through an osteoclast lineage.31,32 Similarly, miR-34a-overexpressing transgenic mice exhibited lower bone resorption and higher bone mass, whereas miR-34a knockout and heterozygous mice displayed elevated bone resorption and reduced bone mass.33
As regulation of bone homeostasis depends on a carefully orchestrated crosstalk among bone-residing cells, miRNAs that are capable of regulating differentiation and/or activity of any bone stromal cells could markedly influence overall bone deposition and degradation. One such miRNA, miR-34c, is an essential regulator of Notch signaling in osteoblasts by directly targeting Notch1, Notch2 and Jagged1, as well as Satb2 and Runx2.34 Interestingly, miR-34c has been shown to function with both cell- and non-cell-autonomous activities. Upregulation of miR-34c inhibits osteogenesis by decreasing Satb2 and Runx2 in osteoblasts, whereas inhibition of Notch signaling may regulate the RANKL/OPG ratio, leading to increased osteoclast differentiation.34,35
Given the profound effect that regulated miRNA expression has on bone marrow cell differentiation and activity, it is perhaps not surprising that misregulation of miRNAs has also been linked to a number of bone-related pathologies, including osteoporosis36,37,38 and osteosarcoma.39,40,41,42,43 Importantly, misregulated miRNAs have been proposed as potential biomarkers of the specific bone-related diseases as they may accurately reflect the disease state of the bone. Because osteoporosis is caused by an imbalance between bone formation and resorption, misregulated bone marrow miRNAs represent promising indicators of assessment of bone fragility and provide potential therapeutic options to restore homeostasis. To this end, several circulating miRNAs (miR-21, miR-23a, miR-24, miR-25, miR-100, miR-125b and miR-133a), most of which have previously been linked to the regulation of bone metastasis (Table 1), have emerged as novel biomarkers of bone turnover in osteoporosis,44,45 demonstrating the feasibility of using miRNAs as biomarkers of bone disease.
Bone-derived miRNAs in bone metastasis
In addition to mediating bone development and homeostasis under physiological conditions, miRNAs can influence essential tumor–stroma interactions that mediate metastasis. Many bone metastatic cancers dysregulate tumor-intrinsic miRNA expression to establish these metastasis-promoting tumor–stroma interactions. For instance, miR-33a is downregulated in lung cancer cells because it functions as a bone metastasis suppressor by targeting PTHrP, a known activator of osteolytic bone resorption, and reducing the ability of cancer cells to induce osteoclast differentiation and activity.46 However, it is also possible for aberrant expression of stromal miRNAs to enhance bone metastatic progression. Thus, any of the miRNAs involved in bone formation and homeostasis described above may potentially participate in osteolytic or osteoblastic bone metastasis if their expression was altered by bone metastatic cancer cells. Such dysregulated miRNAs could potentially be used as biomarkers of bone metastatic progression or as therapeutic targets. However, the levels and function of the majority of these miRNAs during bone metastasis has first to be elucidated before their clinical usefulness can be assessed. Here, we review the known miRNAs expressed in bone stromal cells that are tumor-induced and/or influence bone metastasis (Figure 2 and Table 2).
Figure 2.
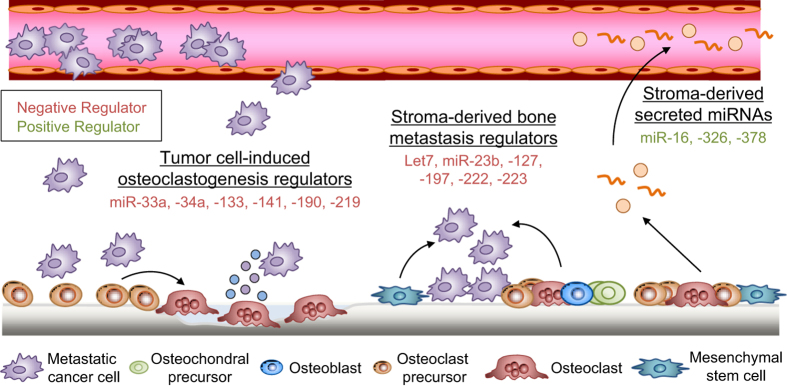
Bone stromal cell microRNA (miRNA) involvement in bone metastasis. MiRNAs regulate bone metastasis activity, including the tumor–stroma crosstalk that mediates osteolysis. Bone metastatic tumor cells promote osteoclastogenesis by repressing a number of negative (red) regulators of osteoclast differentiation. In turn, bone marrow stromal cells use miRNA-mediated regulation to decrease proliferation and mobility and induce dormancy in bone metastatic cells. The modified bone microenvironment releases cell-free and exosomal miRNAs that can be detected in the serum of cancer patients.
Table 2. MiRNAs in bone metastasis.
| miRNA | Cell type | Activity | Species | Cancer type | Reference |
|---|---|---|---|---|---|
| Biomarkers of bone metastasis | |||||
| MiR-16 | Osteoclasts | Increased in presence of bone metastases | Human and mouse | Breast | 47 |
| MiR-326 | Osteoclasts | Increased in presence of bone metastases | Mouse | Lung | 57 |
| MiR-378 | Osteoclasts | Increased in presence of bone metastases | Human and mouse | Breast | 47 |
| miRNA | Cell type | Activity | Targets | Cancer type | Reference |
|---|---|---|---|---|---|
| Regulators of bone metastasis | |||||
| Let-7 | BM-MSC | Decreases mobility of cancer cells | IL-6 | Prostate | 58 |
| MiR-23b | BM-MSC | Decreases proliferation and mobility of cancer cells | MARCKS | Breast | 60 |
| MiR-33a | Osteoclasts | Decreases osteoclastogenesis | PTHrP | Breast and lung | 41,47 |
| MiR-34a | Osteoclasts | Decreases bone metastasis in transgenic mouse model | Tgif2 | Breast and skin | 33 |
| MiR-127 | BM stromal cells | Decreases proliferation of cancer cells, induced dormancy | CXCL12 | Breast | 59 |
| MiR-133a | Osteoclasts | Decreases osteoclastogenesis | Mitf, Mmp14 | Breast | 47 |
| MiR-141 | Osteoclasts | Decreases osteoclastogenesis, inhibits bone metastasis in vivo | Mitf, Calcr | Breast | 47 |
| MiR-190 | Osteoclasts | Decreases osteoclastogenesis | Calcr | Breast | 47 |
| MiR-197 | BM stromal cells | Decreases proliferation of cancer cells, induced dormancy | CXCL12 | Breast | 59 |
| MiR-219 | Osteoclasts | Decreases osteoclastogenesis, inhibits bone metastasis in vivo | Mitf, Traf6 | Breast | 47 |
| MiR-222 | BM stromal cells | Decreases proliferation of cancer cells, induced dormancy | CXCL12 | Breast | 59 |
| MiR-223 | BM stromal cells | Decreases proliferation of cancer cells, induced dormancy | CXCL12 | Breast | 59 |
Abbreviations: BM, bone marrow; miR, microRNA; MSC, mesenchymal stem cell.
During osteolytic metastasis, tumor cells co-opt the regulatory signaling pathways within bone marrow cells to recruit and activate osteoclasts, which in turn orchestrate bone degradation. Any miRNAs that functionally promote or inhibit osteoclastogenesis during this process could represent a good biomarker for osteolytic bone metastasis or serve as a therapeutic target to diminish bone metastasis. One such miRNA, miR-34a, has been shown to decrease osteoclastogenesis and bone resorption under physiological conditions and increase bone mass in transgenic mice, which specifically overexpress miR-34a in osteoclasts.33 MiR-34a, which is downregulated during osteoclast differentiation, could diminish osteoporosis as well as bone metastasis of breast and skin cancers in the same osteoclastic miR-34a transgenic mice model by directly inhibiting the expression of Tgif2.33 Hence, a key osteoclast suppressor miRNA involved in normal bone turnover could potentially become a therapeutic target to confer skeletal protection and ameliorate bone metastasis of cancers.
Recently, an miRNA signature induced by highly metastatic tumor cells that stimulates differentiation of osteoclasts and recruits preosteoclasts to the site of the tumor–bone interface has been described.47 Several miRNAs within the signature have been shown to directly influence osteoclast activity during bone metastasis, including miR-33a, miR-133a, miR-141, miR-190 and miR-219.47 Interestingly, similar to miR-34a, in vitro osteoclastogenesis assays revealed a necessity for these miRNAs to be downregulated. Their ectopic expression, on the other hand, inhibited differentiation through the functional targeting of essential osteoclast genes acting at different stages of osteoclast differentiation. Specifically, miR133a, miR-141 and miR-219 were all found to repress expression of Mitf. In addition, miR-133a was found to repress Mmp14, miR-141 and miR-190 repressed Calcr and miR-219 was able to repress Traf6.47 Importantly, systemic inoculation of pre-miRNA oligonucleotides for miR-141 or miR-219 was sufficient to inhibit bone metastasis from human breast cancer cells.47 Thus, it is possible for bone marrow cell miRNAs to be used as biomarkers of tumor-induced bone microenvironmental changes and osteolytic bone metastasis, as well as serve as potential therapeutic targets.
In addition to the biological significance of intracellular miRNAs, recent studies revealed the functional importance of secreted miRNAs.48,49,50 miRNAs can be released from cells as cell-free miRNAs either bound to protein, such as AGO2, or lipid carriers, such as HDL or LDL, or packaged into microvesicles such as exosomes (30–100 nm) or larger microparticles (100–1000, nm).51 The presence of their protein- or lipid-based carriers or their encapsulation into vesicles confer circulating miRNAs remarkable stability under a variety of harsh environmental conditions.50,51 As such, secreted miRNAs represent important potential biomarkers for diagnosis and prognosis that can be detected by noninvasive means in a reliable and reproducible manner. In fact, several studies have reported the use of circulating miRNAs as biomarkers to predict tumor progression and the presence of bone metastases.51,52,53,54 Furthermore, highly expressed circulating miRNAs from cancer patients have been reported to return to a normal level after tumor resection,55,56 suggesting that the level of circulating miRNAs reflects the disease state of the patient to some extent and could be used as a pharmacodynamic marker. Moreover, cell-free and exosomal miRNAs are of particular interest owing to their pleiotropic roles in modulating many physiological and pathological processes, including cancer metastasis.48
Serum miRNAs shown to be upregulated during metastasis-induced osteoclast differentiation could thus be used as an indicator of osseous metastases or as a potential therapeutic strategy. Two such osteoclastic miRNAs, miR-16 and miR-378, were found significantly elevated in the serum from mice bearing highly metastatic breast cancer cells, and in bone lesions and serum from patients with breast cancer metastatic to bone as compared with healthy female donors.47 In fact, miR-16 and miR-378 possessed comparable sensitivity relative to the bone turnover marker N-terminal telopeptide (NTX), indicating the potential for using these miRNAs as biomarkers for bone metastasis progression. In a separate study, serum levels of miR-326 strongly associated with tumor burden and a marker of bone metastatic burden, PINI (procollagen I amino-terminal propeptide), in a mouse model of lung cancer bone metastasis.57 These results indicate that miRNAs such as miR-16, miR-326 and miR-378 could potentially serve as novel biochemical markers for monitoring osteolytic bone metastatic progression.
Bone stromal miRNAs are also capable of directly influencing metastatic cells. Let-7c inhibited IL-6 within bone marrow MSCs (BM-MSCs), whereas ectopic expression of Let-7c was capable of repressing the migration- and invasion-promoting potential of tumor-associated BM-MSCs.58 A separate study found that several miRNAs were transferred from bone marrow stromal cells to nearby breast cancer cells through gap junctions and to a smaller extent via exosomes upon seeding to the bone.59 MiR-127, miR-197, miR-222 and miR-223, all direct inhibitors of CXCL12, inhibited proliferation and arrested the metastatic cells in a dormant state in which the latter could survive high-dose chemotherapy, making these miRNAs interesting targets for combination treatment with conventional therapy in which dormancy is an obstacle. Moreover, exosomal miR-23b has also recently been shown to induce dormancy in breast cancer cells after being released by BM-MSCs.60 Furthermore, transfer of stromal exosomes containing miR-23b was shown to inhibit cell cycling and mobility by targeting MARCKS in cancer cells. Interestingly, metastatic breast cancer cells in patient bone marrow had increased miR-23b and decreased MARCKS expression. Taken together, some bone stromal miRNAs may be useful biomarkers for specific metastasis-related processes such as increased chemoresistance or quiescence, which could direct the therapeutic regimen.
miRNAs as markers and therapeutic targets
It is currently difficult to accurately diagnose patients with emerging bone metastasis, and standard biomarkers, such as NTX, show only modest specificity as diagnostic markers. miRNAs expressed by the bone stroma could lead to improvements in diagnosis, treatment and prevention of bone metastases and elucidate the unique aspect of the bone microenvironment that supports tumor growth in the bone. More specifically, characterization of expression and secretion of bone marrow-derived miRNAs could enable the selection of patients at increased risk of bone metastasis for appropriate targeted therapies. Such patient selection may be achieved by simple quantitative diagnostic tests, such as quantitative reverse transcription-PCR, for specific miRNAs whose levels correlate with bone metastatic progression. Such diagnostic tests would probably analyze miRNA levels of a clinical ‘bone metastatic signature, which is likely to be made up of miRNAs specifically expressed by metastatic or bone metastatic cancer cells, as well as miRNAs whose expression is altered owing to tumor-induced changes in the bone microenvironment. Moreover, such miRNA-based diagnostic tests will likely be combined with other diagnostic and prognostic biomarkers of cancer progression or bone metastasis (such as serum proteins) to increase sensitivity and specificity of detection. This diagnostic approach would identify cancer patients at high risk for bone metastasis, therapeutic resistance or dormant cancer cells in the bone, who would greatly benefit from a preventive treatment or initiate therapies at an earlier state of metastatic lesion. Moreover, owing to their potential use as pharmacodynamic biomarkers whose levels depict the disease state of the patient, miRNAs possess the potential to guide treatment regimens during cancer therapy.
Furthermore, miRNAs with functional roles in promoting bone metastatic progression could become important therapeutic targets. Potential therapies for such metastasis-promoting miRNAs include siRNA or antisense oligonucleotides, miRNA sponges, miRNA masking and small-molecule inhibitors. Furthermore, endogenous or synthetic exosomes could be used to efficiently deliver these or other inhibitors in a tissue-specific manner.61,62 On the other hand, for metastasis-inhibiting miRNAs usually downregulated during cancer progression, administration of synthetic miRNAs or forced expression of endogenous miRNAs may be a useful therapeutic option to restore expression. Using systemic inoculation of pre-miRNA oligonucleotides for osteoclastic miRNAs miR-141 or miR-219, Ell et al.47 successfully inhibited bone metastasis from human breast cancer cells. As intravenously injected exosomes are detectable in the bone marrow of mice,63 miRNAs could also be delivered using exosomes. However, the development of miRNA-based therapeutics will require a more complete understanding of miRNA–mRNA targeting in diverse tissue types to minimize potential side effects owing to the multitarget nature of miRNAs in gene regulation. Moreover, identifying the specific targets through which these miRNAs exert their function in bone metastasis also opens possibilities to identify potential downstream targets of therapeutic interest.
Conclusions
miRNA activity represents an essential level of gene regulation during skeletal development and homeostasis. Ablation of miRNA activity, through the inhibition of miRNA processing pathway components such as Dicer or AGO2, has been shown to inhibit the development of multiple bone marrow stromal cells, resulting in devastating skeletal defects. In addition, dysregulation of individual miRNAs inhibited the differentiation and activity of multiple cell types and precipitated pathological events. Importantly, miRNA regulation has also been shown to have pivotal roles in regulating the development and progression of bone metastasis. In particular, the role of bone stromal miRNAs, such as those expressed in osteoclasts, as mediators or inhibitors of bone metastasis and the changes in their expression levels during disease progression have only recently started to be uncovered. Such knowledge, however, will be vital in further deepening our understanding of bone metastatic progression and transferring the knowledge from the bench into the clinical setting.
Bone marrow stroma-derived miRNAs constitute a novel class of dysregulated molecules that could provide new avenues for diagnosis and treatment of bone metastases. In particular, circulating miRNAs derived from tumor-induced changes in the bone microenvironment represent a novel class of biomarkers with large diagnostic and prognostic potential. This potential is based on their noninvasive detection in body fluids and their high resistance and stability under various conditions that could degrade the majority of RNAs. In addition, several methodologies are available for establishing miRNA expression levels and signatures in a high-throughput and robust manner. Although the recent data are exclusively preclinical evidence, the application of bone marrow stroma-derived miRNAs in metastasis therapy as adjuvant tools or targets appears very promising. A better understanding of the complex network of genes and cellular signaling transduction pathways regulated by miRNAs would enrich our knowledge of metastatic progression and hence improve the therapeutic outcome of cancer patients.
Acknowledgments
Research in our laboratory was supported by grants from the Komen for the Cure (KG110464), Breast Cancer Research Foundation, Department of Defense (BC123187), Brewster Foundation and the National Institutes of Health (R01CA134519 and R01CA141062) (to YK).
Footnotes
The authors declare no conflict of interest.
References
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006; 1: R17–R29. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11: 597–610. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012; 13: 271–282. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science 2011; 331: 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004; 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003; 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003; 423: 337–342. [DOI] [PubMed] [Google Scholar]
- Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature 2003; 423: 349–355. [DOI] [PubMed] [Google Scholar]
- Ell B, Kang Y. MicroRNAs as regulators of bone homeostasis and bone metastasis. BoneKEy Rep 2014; 3: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol 2010; 340: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA 2008; 105: 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem 2009; 284: 4667–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood 2011; 117: 3648–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T et al. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem 2010; 109: 866–875. [DOI] [PubMed] [Google Scholar]
- Sugatani T, Hildreth BE 3rd, Toribio RE, Malluche HH, Hruska KA. Expression of DGCR8-dependent microRNAs is indispensable for osteoclastic development and bone-resorbing activity. J Cell Biochem 2014; 115: 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet 2008; 40: 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol 2011; 31: 3019–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai X-H, Luan Y, Liu C-J. miR-199a*, a bone morphogenic protein 2-responsive microRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem 2009; 284: 11326–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One 2011; 6: e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xie R-l, Croce CM, Stein JL, Lian JB, van Wijnen AJ et al. . A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci 2011; 108: 9863–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Gordon JAR, Beloti MM, Croce CM, van Wijnen AJ, Stein JL et al. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci USA 2010; 107: 19879–19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA 2009; 106: 20794–20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA 2011; 108: 6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem 2007; 101: 996–999. [DOI] [PubMed] [Google Scholar]
- Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E et al. MiR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol 2013; 228: 1506–1515. [DOI] [PubMed] [Google Scholar]
- Franceschetti T, Kessler CB, Lee SK, Delany AM. MiR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem 2013; 288: 33347–33360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi F, Murakami Y, Saito T, Miyasaka N, Kohsaka H. MiR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res Ther 2013; 15: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cheng P, Xie H, Zhou HD, Wu XP, Liao EY et al. MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res 2014; 29: 338–347. [DOI] [PubMed] [Google Scholar]
- Guo LJ, Liao L, Yang L, Li Y, Jiang TJ. MiR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp Cell Res 2014; 321: 142–152. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W et al. Interferon-beta-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett 2012; 586: 3255–3262. [DOI] [PubMed] [Google Scholar]
- Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci USA 2010; 107: 15804–15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang T-C et al. MiR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 2014; 512: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T et al. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet 2012; 21: 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 2008; 14: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F-S, Chung P-C, Lin C-L, Chen M-W, Ke H-J, Chang Y-H et al. MicroRNA-29a protects against glucocorticoid-induced bone loss and fragility in rats by orchestrating bone acquisition and resorption. Arthritis Rheumat 2013; 65: 1530–1540. [DOI] [PubMed] [Google Scholar]
- He X, Zhang W, Liao L, Fu X, Yu Q, Jin Y. Identification and characterization of microRNAs by high through-put sequencing in mesenchymal stem cells and bone tissue from mice of age-related osteoporosis. PLoS One 2013; 8: e71895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xie H, Liu W, Hu R, Huang B, Tan Y-F et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest 2009; 119: 3666–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyan W, Shuhua Y, Xiufang W, Xiaoyun L. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med Oncol 2011; 28: 1469–1474. [DOI] [PubMed] [Google Scholar]
- Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y et al. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther 2011; 19: 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KB, Salah Z, Sara DM, Galasso M, Gaudio E, Nuovo GJ et al. MicroRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res 2012; 72: 1865–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hou C, Zhang H, Wang D, Ma Y, Zhang Y et al. MiR-126 functions as a tumor suppressor in osteosarcoma by targeting Sox2. Int J Mol Sci 2014; 15: 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X et al. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting ezrin. Am J Pathol 2012; 180: 2440–2451. [DOI] [PubMed] [Google Scholar]
- Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS et al. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res 2014; 29: 1718–1728. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Moore BT, Peng X-H, Fang X, Lappe JM et al. MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One 2012; 7: e34641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PL, Liao SH, Hung JY, Huang MS, Hsu YL. MicroRNA-33a functions as a bone metastasis suppressor in lung cancer by targeting parathyroid hormone related protein. Biochim Biophys Acta 2013; 1830: 3756–3766. [DOI] [PubMed] [Google Scholar]
- Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 2013; 24: 542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liang H, Zhang J, Zen K, Zhang C-Y. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol 2012; 22: 125–132. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–659. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014; 11: 145–156. [DOI] [PubMed] [Google Scholar]
- Alečković M, Kang Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim Biophys Acta 2014; 1855: 24–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururajan M, Josson S, Chu GC-Y Lu C-L, Lu Y-T, Haga CL et al. MiR-154* and miR-379 in the DLK1-DIO3 microrna mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin Cancer Res 2014; 20: 6559–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Wang Z, Zhang R, Feng W. High serum microRNA-335 level predicts aggressive tumor progression and unfavorable prognosis in pediatric acute myeloid leukemia. Clin Transl Oncol (e-pub ahead of print 10 October 2014). [DOI] [PubMed] [Google Scholar]
- Zhao FL, Hu GD, Wang XF, Zhang XH, Zhang YK, Yu ZS. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J Int Med Res 2012; 40: 859–866. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009; 14: 529–538. [DOI] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating micrornas as novel minimally invasive biomarkers for breast cancer. Ann Surg 2010; 251: 499–505. [DOI] [PubMed] [Google Scholar]
- Valencia K, Martín-Fernández M, Zandueta C, Ormazábal C, Martínez-Canarias S, Bandrés E et al. MiR-326 associates with biochemical markers of bone turnover in lung cancer bone metastasis. Bone 2013; 52: 532–539. [DOI] [PubMed] [Google Scholar]
- Sung SY, Liao CH, Wu HP, Hsiao WC, Wu IH, Yu J et al. Loss of let-7 microRNA upregulates IL-6 in bone marrow-derived mesenchymal stem cells triggering a reactive stromal response to prostate cancer. PLoS One 2013; 8: e71637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 2011; 71: 1550–1560. [DOI] [PubMed] [Google Scholar]
- Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi RU et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Science Signal 2014; 7: ra63. [DOI] [PubMed] [Google Scholar]
- Kooijmans SAA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomed 2012; 7: 1525–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhal S, Wood MJ. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. BioEssays 2011; 33: 737–741. [DOI] [PubMed] [Google Scholar]
- Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum 2011; 63: 1582–1590. [DOI] [PubMed] [Google Scholar]
- Cheng P, Chen C, He H, Hu R, Zhou H, Xie H et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res 2013; 28: 1180–1190. [DOI] [PubMed] [Google Scholar]
- Vimalraj S, Partridge NC, Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol 2014; 229: 1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Ma J, Chen S, Chen X, Yu X. MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine 2014; 45: 302–310. [DOI] [PubMed] [Google Scholar]
- Li H, Li T, Wang S, Wei J, Fan J, Li J et al. MiR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res 2013; 10: 313–324. [DOI] [PubMed] [Google Scholar]
- Zhang JF, Fu WM, Ml He, Xie WD, Lv Q, Wan G et al. miRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol 2011; 8: 829–838. [DOI] [PubMed] [Google Scholar]
- Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem 2009; 108: 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 2009; 284: 15676–15684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinas K, Kessler CB, Ricks T, Gronowicz G, Delany AM. MiR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 2010; 285: 25221–25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Cheng P, Chen C, He H-B, Xie G-Q, Zhou H-D et al. MiR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res 2012; 27: 1598–1606. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Qu X, Li H, Huang S, Wang S, Xu Q et al. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett 2012; 586: 2375–2381. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T et al. MiR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun 2008; 368: 267–272. [DOI] [PubMed] [Google Scholar]
- Goettsch C, Rauner M, Pacyna N, Hempel U, Bornstein SR, Hofbauer LC. MiR-125b regulates calcification of vascular smooth muscle cells. Am J Pathol 2011; 179: 1594–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Nozawa Y, Y Akao. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem 2009; 284: 19272–19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Zhang J, Yuan T, Ma B. MiR-143 suppresses osteogenic differentiation by targeting Osterix. Mol Cell Biochem 2014; 390: 69–74. [DOI] [PubMed] [Google Scholar]
- Bhushan R, Grünhagen J, Becker J, Robinson PN, Ott C-E, Knaus P. MiR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int J Biochem Cell Biol 2013; 45: 696–705. [DOI] [PubMed] [Google Scholar]
- Kim KM, Park SJ, Jung S-H, Kim EJ, Jogeswar G, Ajita J et al. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 2012; 27: 1669–1679. [DOI] [PubMed] [Google Scholar]
- Itoh T, Takeda S, Akao Y. MicroRNA-208 modulates BMP-2-stimulated mouse preosteoblast differentiation by directly targeting V-ets erythroblastosis virus E26 oncogene homolog 1. J Biol Chem 2010; 285: 27745–27752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A et al. MiR-214 targets ATF4 to inhibit bone formation. Nat Med 2013; 19: 93–100. [DOI] [PubMed] [Google Scholar]
- Shi K, Lu J, Zhao Y, Wang L, Li J, Qi B et al. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone 2013; 55: 487–494. [DOI] [PubMed] [Google Scholar]
- Gamez B, Rodriguez-Carballo E, Bartrons R, Jl Rosa, Ventura F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J Biol Chem 2013; 288: 14264–14275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res 2011; 26: 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureel J, Dixit M, Tyagi AM, Mansoori MN, Srivastava K, Raghuvanshi A et al. MiR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis 2014; 5: e1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Fu WM, Ml He, Wang H, Wang WM, Yu SC et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell 2011; 22: 3955–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisse TS, Chun RF, Rieger S, Adams JS, Hewison M. Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts. J Bone Miner Res 2013; 28: 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Ren F, Wang Y, Li S, Gao Z, Wang X et al. MiR-764-5p promotes osteoblast differentiation through inhibition of CHIP/STUB1 expression. J Bone Miner Res 2012; 27: 1607–1618. [DOI] [PubMed] [Google Scholar]
- Papaioannou G, Inloes JB, Nakamura Y, Paltrinieri E, Kobayashi T. Let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc Natl Acad Sci USA 2013; 110: E3291–E3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S et al. Germline deletion of the miR-17[sim]92 cluster causes skeletal and growth defects in humans. Nat Genet 2011; 43: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Song J, Kim S, Park HM, Chun CH, Sonn J et al. MicroRNA-34a modulates cytoskeletal dynamics through regulating RhoA/Rac1 cross-talk in chondroblasts. J Biol Chem 2012; 287: 12501–12509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Abouheif MM, Nakasa T, Shibuya H, Niimoto T, Kongcharoensombat W, Ochi M. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology 2010; 49: 2054–2060. [DOI] [PubMed] [Google Scholar]
- Kim D, Song J, Jin EJ. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J Biol Chem 2010; 285: 26900–26907. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhong N, Sun J, Min Z, Zhao W, Zhang R, Wang W et al. MicroRNA-337 is associated with chondrogenesis through regulating TGFBR2 expression. Osteoarthritis Cartilage 2012; 20: 593–602. [DOI] [PubMed] [Google Scholar]
- Paik S, Jung HS, Lee S, Yoon DS, Park MS, Lee JW. MiR-449a regulates the chondrogenesis of human mesenchymal stem cells through direct targeting of lymphoid enhancer-binding factor-1. Stem Cells Dev 2012; 21: 3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Murphy CL. MiR-1247 functions by targeting cartilage transcription factor SOX9. J Biol Chem 2013; 288: 30802–30814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-B, Zhong W-J, Wang L. A signal-amplification circuit between miR-218 and Wnt/β-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone 2014; 58: 59–66. [DOI] [PubMed] [Google Scholar]