Figure 2. c-NHEJ generates chromosomal translocations in HCT116 human cells.
A. Induction of chromosomal translocations with sequence-specific nucleases. Derivative chromosomes. Der19 and Der22 were detected by PCR using primers that flank the cleavage sites after ZFNp84 and ZFNEWS expression.
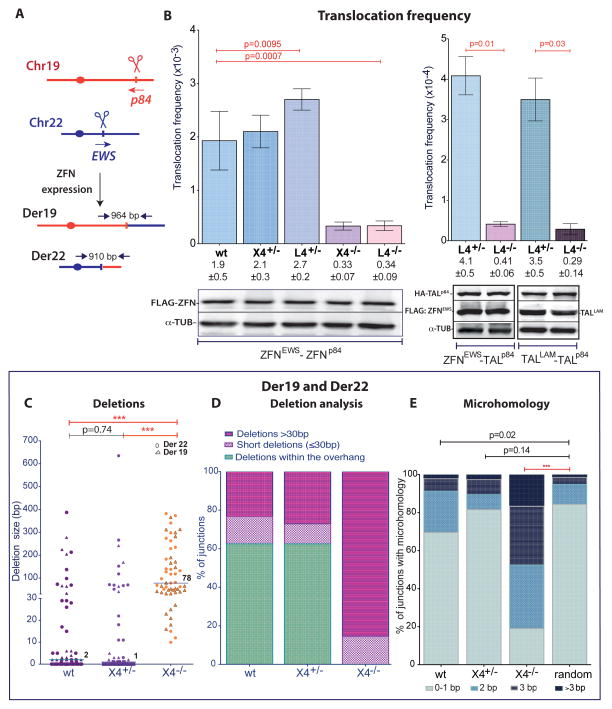
B. Translocation frequency is reduced in L4−/− and X4−/− HCT116 cells. Translocations were quantified as follows: ZFNEWS and ZFNp84, Der19 and Der22 (wt, n = 7; L4, n = 3; X4, n = 4); ZFNEWS and TALp84, Der22 (n = 4); TALLAM and TALp84, Der1 (n = 4). Der19 and Der22 were assessed in the same experiment and the frequencies averaged. Error bars, +/− SEM. Nuclease expression was detected 48 h after transfection.
C. – E. Translocation junction analysis from X4−/− cells demonstrates a shift towards longer deletions and an increased presence of microhomology. Der19 and Der22 junctions derived from ZFN expression were pooled. C, Deletion lengths from individual Der19 and Der22 junctions are indicated by the triangle and circle, respectively. D, Junctions are grouped according to whether the deletions were restricted to the overhang or were short (≤ 30 bp) or long (> 30 bp) deletions extending outside of the overhang. E, Microhomology distribution.
See also Figure S2.