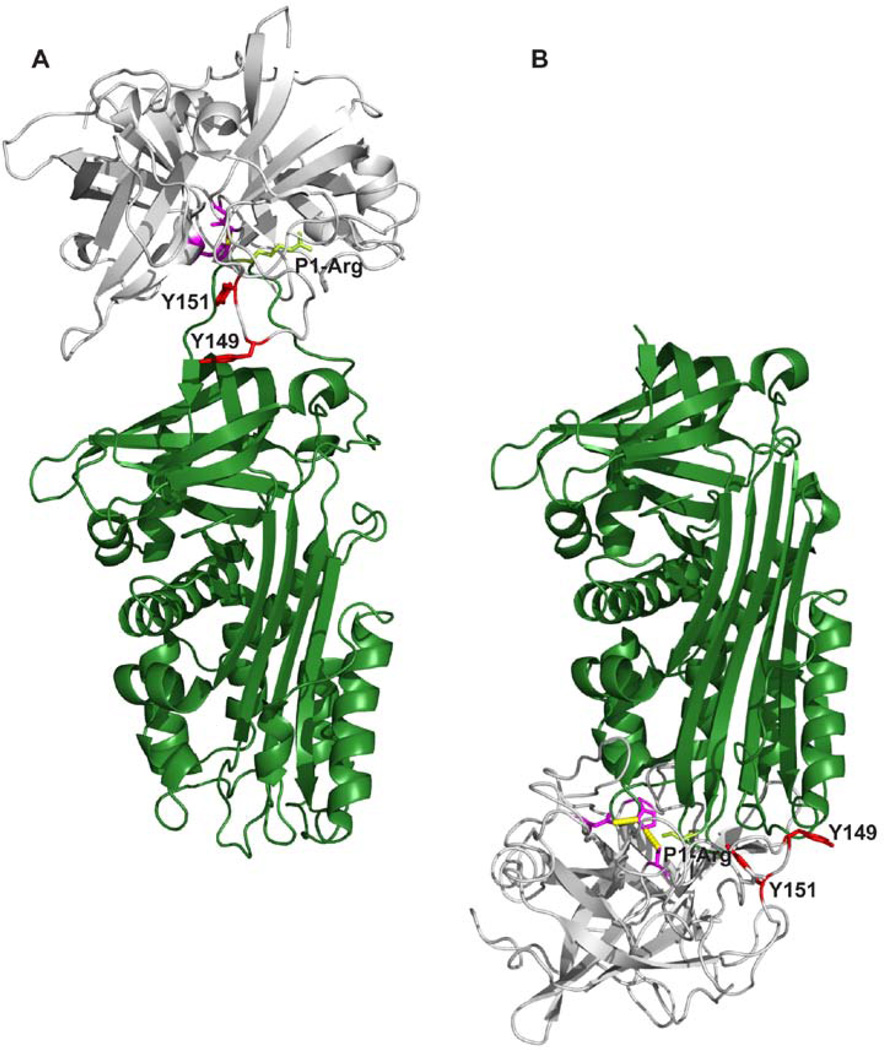
Figure 10. A model of the complexes between uPA and PAI-1.
A, The Michaelis complex between uPA and PAI-1, with uPA in gray and PAI-1 in dark green [39]. The catalytic triad of uPA is coloured purble with the two tyrosine residues, Y149 and Y151, involved in the epitopes of the conformation-sensitive antibodies depicted in red. Shown in light green are the side chain of the P1-arginine residue of PAI-1. B, A model of the stable uPA-PAI-1 complex build on the basis of the complex between α1-antitrypsin and trypsin [45], with the same colour codes as in A. All figures were constructed with Pymol [41] on the basis of the coordinates given in the PDB entry 3CVM for PAI-1 [46], 1LMW for two-chain active uPA [47], 1EZX for the stable covalent complex between α1-antitrypsin and trypsin [45] and 3PB1 for the Michaelis complex between uPA and PAI-1[39].