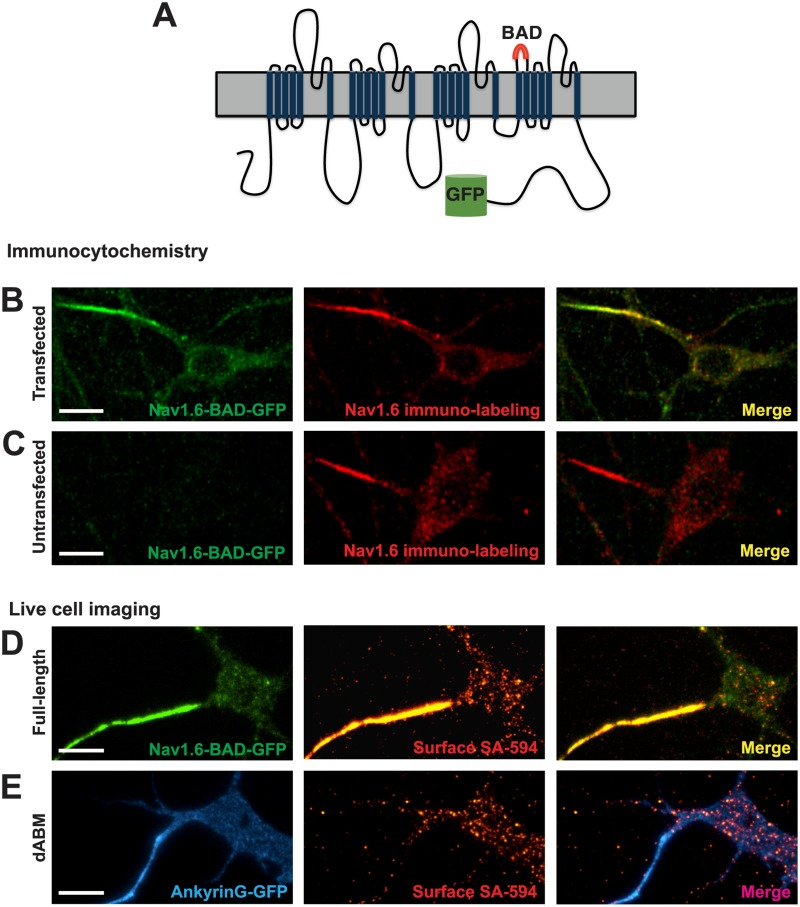
Fig 1. Nav1.6-BAD-GFP expression mimics that of the endogenous channel in cultured rat hippocampal neurons.
A) Schematic of the Nav1.6-BAD-GFP construct showing location of the biotin acceptor domain (BAD) and fusion of GFP to the channel C-terminus. (B-C) Comparison of anti-Nav1.6 immunolabeling in transfected and untransfected hippocampal neurons. Compressed confocal z-stack of GFP fluorescence (left panel) and anti-Nav1.6 immunolabeling (middle panel) from a Nav1.6-BAD-GFP transfected neuron (B) or a non-transfected neuron (C). Overlay (right panel). Fluorescence intensity of immunolabeling for transfected vs endogenous AIS Nav1.6 expression was not significant (t-test, p = 0.132). (D-E) Surface expression of full-length Nav1.6 or Nav1.6 lacking the ankG-binding motif (ABM) in live neurons visualized by TIRF microscopy. D) Nav1.6-BAD-GFP is highly enriched at the AIS as indicated both by total expressed protein (green) and surface expression (red) visualized by live cell labeling with SA-594. E) Nav1.6-dABM-BAD co-expressed with ankG-GFP (blue) as an axonal marker. Surface channels labeled with SA-594 (red) demonstrated a lack of axonal localization while maintaining a somatic expression pattern similar to the full-length channel. Panels B and C are taken from DIV12 cultures since the endogenous Nav1.6 does not appear until this developmental time. Panels D and E are from DIV8 cultures. Scale bars represent 10 μm.