Abstract
The bacterial flagellar motor has a stator and a rotor. The stator is composed of two membrane proteins, MotA and MotB in Escherichia coli and PomA and PomB in Vibrio alginolyticus. The Vibrio motor has a unique structure, the T ring, which is composed of MotX and MotY. Based on the structural information of PomB and MotB, we constructed three chimeric proteins between PomB and MotB, named PotB91, PotB129, and PotB138, with various chimeric junctions. When those chimeric proteins were produced with PomA in a ΔmotAB strain of E. coli or in ΔpomAB and ΔpomAB ΔmotX strains of Vibrio, all chimeras were functional in E. coli or Vibrio, either with or without the T ring, although the motilities were very weak in E. coli. Furthermore, we could isolate some suppressors in E. coli and identified the mutation sites on PomA or the chimeric B subunit. The weak function of chimeric PotBs in E. coli is derived mainly from the defect in the rotational switching of the flagellar motor. In addition, comparing the motilities of chimera strains in ΔpomAB, PotB138 had the highest motility. The difference between the origin of the α1 and α2 helices, E. coli MotB or Vibro PomB, seems to be important for motility in E. coli and especially in Vibrio.
Keywords: Basal body, flagellar motor, T ring, V. alginolyticus
Introduction
Many bacteria can swim using flagella. Each flagellum consists of a filament, a hook, and a basal body. The rotation of a flagellum is made by the interaction between the rotor and the stator (Morimoto and Minamino 2014). The rotor is called the C ring, which is associated beneath the basal body, and is composed of FliG, FliM, and FliN. Multiple stator units assemble around each rotor. There are two types of flagellar motors depending on the coupling ion. One is a proton driven motor with a stator composed of MotA and MotB, and is found in bacteria such as Escherichia coli or Salmonella. The other is a Na+ driven motor with a stator composed of PomA and PomB, and is found in such bacteria as Vibrio alginolyticus (Yorimitsu and Homma 2001; Blair 2003; Li et al. 2011b). MotA and PomA are four transmembrane (TM) domain proteins and MotB and PomB are one TM domain proteins (Chun and Parkinson 1988; Asai et al. 1997). PomAB or MotAB form heterohexameric channel complexes with an A4:B2 stoichiometry to conduct sodium ions or protons (Sato and Homma 2000; Kojima and Blair 2004; Takekawa et al. 2013). The stator A subunit has a cytoplasmic loop between TM3 and TM4 and charged residues in this loop interact with the C ring component FliG (Zhou et al. 1998b; Morimoto et al. 2010, 2013; Takekawa et al. 2014). The negative charged residue, D24 of PomB or D32 of MotB, is critical for the force generation and constitutes the ion-binding site in the stator channel (Zhou et al. 1998b; Sudo et al. 2009; Terashima et al. 2010b). It has been reported that a specific region, residues 44–58 in Vibrio PomB and 52–65 in E. coli MotB, serves as a plug, which regulates ion influx (Hosking et al. 2006; Kojima et al. 2009; Li et al. 2011a). Torque is generated by the interaction between the stator component PomA (MotA) and the rotor component FliG (Zhou et al. 1998a; Yakushi et al. 2006).
In E. coli and in Salmonella, the basal body is composed of the rod and several ring structures, the L, P, MS, and C rings (Macnab 1992). In V. alginolyticus, the basal body has a T ring and an H ring in addition to the rings found in E. coli (Terashima et al. 2006, 2010a). The T ring is composed of MotX and MotY. When MotX or MotY is deleted, the stator cannot assemble around the rotor or the basal body, indicating that the T ring is required for stator incorporation into the motor (Terashima et al. 2006). MotX affects the membrane localization of PomB, suggesting that PomB interacts with MotX, however, their direct binding has not yet been detected (Okabe et al. 2005). Some crystal structures of fragments of the stator B subunits from Helicobacter pylori and Salmonella have been resolved (Roujeinikova 2008; Kojima et al. 2009). The crystal structure of the periplasmic region of PomB (PomBC) has been recently resolved (Zhu et al. 2014). The N-terminus of PomBC contains six negatively charged residues on the α1 and α2 helices and it forms a negatively charged surface. Interestingly, such a negative surface is not observed in the MotBC structure of Salmonella (Kojima et al. 2009). The isoelectric point (pI) of MotX has been estimated at 8.48, raising the possibility that these negatively charged residues of PomBC are involved in the interaction between PomB and MotX. Note that the estimated pI values of MotX proteins vary among bacteria, suggesting that MotX does not always have a positive charge. The charged residues might cause electrostatic interactions between these helices of PomB and MotX. To test this idea, we made charge reversal point mutants in the α1 helix or α2 helix of PomBC. The single mutations or the quintuple mutation conferred a swimming ability similar to the wild-type protein when induced by 0.02% arabinose, suggesting that those charge residues of the α1 or α2 helices are not critical for motor function (Zhu et al. 2014). In this study, we made several new chimeric proteins whose junction sites were designed at the N terminus of the α1 helix to the C terminus of the α2 helix. We used those chimeric proteins to determine the specificity of the B subunit of the flagellar motor protein for Vibrio and E. coli.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are shown in Table1. Vibrio alginolyticus was cultured in VC broth [0.5% (w/v) Polypeptone, 0.5% (w/v) Bacto yeast extract, 0.4% (w/v) K2HPO4, 3% (w/v) NaCl, 0.2% (w/v) d-glucose] or in VPG medium [1% (w/v) Polypeptone, 0.4% K2HPO4, 3% (w/v) NaCl, 0.5% glycerol] at 30°C. Escherichia coli was cultured in LB broth [1% (w/v) Bactotryptone, 0.5% (w/v) Bacto yeast extract, 0.5% (w/v) NaCl] at 37°C or TG broth [1% (w/v) Bactotryptone, 0.5% (w/v) NaCl, 0.5% (w/v) glycerol] at 30°C. Chloramphenicol was added to a final concentration of 2.5 μg/mL for V. alginolyticus and 25 μg/mL for E. coli.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | Recipient for cloning experiments | |
| RP6894 | ΔmotAB | J. S. Parkinson |
| Vibrio alginolyticus | ||
| NMB191 | ΔpomAB | Yorimitsu et al. (1999) |
| TH4 | ΔpomAB ΔmotX | Terashima et al. (2006) |
| Plasmid | ||
| pBAD33 | Cmr, PBAD | Guzman et al. (1995) |
| pHFAB | pBAD33/pomA, pomB | Fukuoka et al. (2005) |
| pTF9 | pBAD33/pomA, potB59 | H. Fukuoka |
| pTSK62 | pBAD33/pomA, potB91 | This study |
| pTSK63 | pBAD33/pomA, potB129 | This study |
| pTSK64 | pBAD33/pomA, potB138 | This study |
Cmr, chloramphenicol resistant; PBAD; arabinose promoter.
Swimming assay in soft agar plates
Cells were inoculated in VPG soft agar plates [1% (w/v) Polypeptone, 0.4% K2HPO4, 3% (w/v) NaCl, 0.5% glycerol, 0.25% or 0.3% (w/v) Bacto agar] containing 0.02% (w/v) arabinose or 0.2% (w/v) arabinose and 2.5 μg/mL chloramphenicol for V. alginolyticus or in TB soft agar plates [1% (w/v) Bactotryptone, 0.5% (w/v) NaCl, 0.3% (w/v) Bacto agar] containing 0.02% (w/v) arabinose and 25 μg/mL chloramphenicol for E. coli. The plates were incubated at 30°C for the desired times.
Measurement of swimming speed
Overnight cultures were inoculated into VPG medium for V. alginolyticus and into TG broth for E. coli containing 0.2% (w/v) arabinose or 0.02% (w/v) arabinose at a 100-fold dilution. Cells were grown at 30°C for 4 h for V. alginolyticus and 6 h for E. coli. Swimming of the cells was observed using dark-field microscopy at a 15-fold dilution with the growth medium. Swimming cells were recorded and then analyzed using software for motion analysis (Move-tr/2D; Library Co., Japan) as described previously (Terauchi et al. 2011). Swimming speeds were determined from data of at least 20 individual cells except for PotB138-sp1 (n = 5).
Measurement of switching fraction
The switching fraction was measured by the tethered cell assay. Overnight E. coli cultures were inoculated into TG broth containing 0.02% (w/v) arabinose at a 100-fold dilution. Cells were grown at 30°C for 4 h and washed with 10 mmol/L potassium-phosphate, 10 mmol/L lactic acid, 100 mmol/L NaCl, and 0.1 mmol/L EDTA, pH 7.0. Their flagella were sheared by passing multiple times through a needle syringe. Cells were then attached on glass slides pretreated with an anti-E. coli flagellin antibody (Nishiyama and Kojima 2012). Rotation of the cells was observed using phase-contrast microscopy and was recorded on a computer using Power director (Cyber Link, Japan). Rotational motion of the cells was analyzed using software for motion analysis (Move-tr/2D; Library Co.). The number of switching times was counted in rotating cells for 30 sec. The switching fraction was calculated as the proportion of the number of cells whose flagella showed switching more than once to that of all rotating cells. The switching fraction was determined from about 20 individual cells.
Immunoblotting
Cells were resuspended in sodium dodecyl sulphate (SDS) loading buffer and boiled at 95°C for 5 min, and then samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinyliden fluoride (PVDF) membranes. Immunoblotting was performed as previously described using an anti-Salmonella MotB antibody (Kojima et al. 2008) or an anti-PomA antibody (Yorimitsu et al. 1999). Protein detection was carried out by chemiluminescence using an LAS-3000 (Fujifilm Co., Japan).
Suppressor isolation
Escherichia coli RP6894 (ΔmotAB) carrying plasmids encoding PotB91 (pTSK62), PotB129 (pTSK63) or PotB138 (pTSK64) were freshly inoculated on TB soft agar plates containing 25 μg/mL and 0.02% arabinose. Incubation at 30°C was continued overnight, and the motility halos that emerged from each inoculation were isolated and formed single colonies. After confirmation of the enhanced motility of the isolated suppressors, the plasmids were purified from these strains and were then sequenced to identify the mutation sites. DNA sequencing was performed with an ABI Prism genetic analyzer (Applied Biosystems, Japan).
Results
Motility of Vibrio by the chimeric proteins
We examined the roles of the α1 helix and the α2 helix using chimeric proteins composed of the N-terminus of PomB fused with the C-terminus of MotB. We previously showed that a chimeric protein, PotB, renamed as PotB59, that has the N-terminal region of PomB (1–50) fused to the C-terminal periplasmic region of E. coli MotB (59–308), is functional with PomA in E. coli as well as in strains of V. alginolyticus that are defective in motX or motY (Asai et al. 2003). Based on the structure comparison between PomA and MotB to determine the junction sites (Kojima et al. 2009; Zhu et al. 2014), we made three new chimeric proteins, PotB91, PotB129 and PotB138 (Figs.1, S1). PotB91 consists of PomB (1–135) and MotB (91–308), PotB129, consists of PomB (1–168) and MotB (129–308), and PotB138 consists of PomB (1–175) and MotB (138–308) (Fig.1). We introduced these proteins with PomA into the mutant strains of V. alginolyticus ΔpomAB (NMB191) or ΔpomAB ΔmotX (TH4) and observed their motilities in soft agar plates (Fig.2A and B). All three chimeric proteins conferred motile ability to V. alginolyticus even in the absence of the T ring, however, the original stator complex composed of PomA and PomB did not confer any motility in the absence of MotX or the T ring as previously reported (Asai et al. 2003). The sizes of the swimming rings without MotX or the T ring by the chimeric stators were much smaller than in the T ring containing strain. The sizes of the swimming rings of PotB91, PotB129, and PotB138 were larger than that of PotB59 and the diameter of PotB138 was the largest among the three chimeric proteins. These observations suggest that the α1 helix and especially the α2 helix of PomB are important for function in Vibrio.
Figure 1.
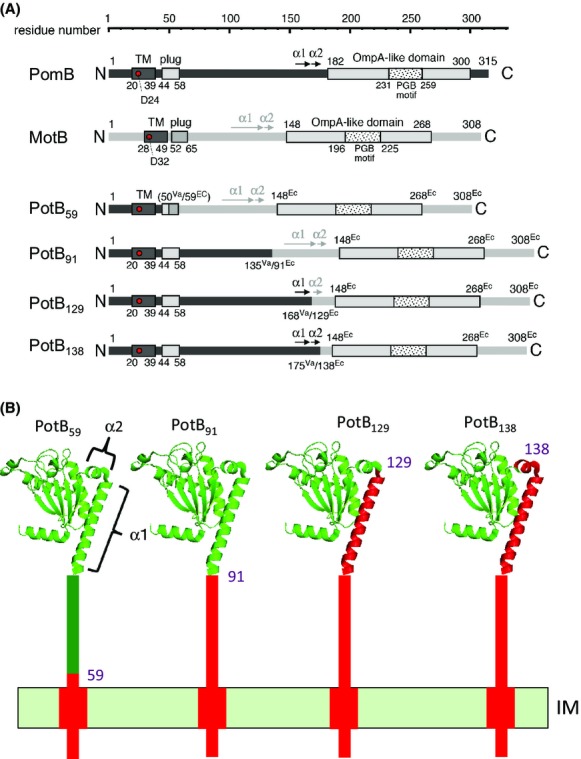
Constructs of the chimeric proteins used in this study. (A) Schematics of the primary structures of PomB, PotB59, PotB91, PotB129 and PotB138. PomB is composed of 315 amino acids and has a single TM domain (residues 20–39) in the N-terminal region, a plug domain (residues 44–58) and a large periplasmic region including an “Omp-A like domain” (residues 182–300). Asp-24, which is essential for ion translocation across the cytoplasmic membrane is shown as a red circle. (B) Topology models of the chimeric proteins. PotB91, PotB129 and PotB138 contain the region prior to the α1 helix of PomB, the region from the N terminal to the α1 helix of PomB, and the region from the N terminal to the α2 helix of PomB, respectively, and the rest of the region is derived from MotB. In the topological models, red indicates regions derived from PomB and green indicates regions derived from MotB.
Figure 2.
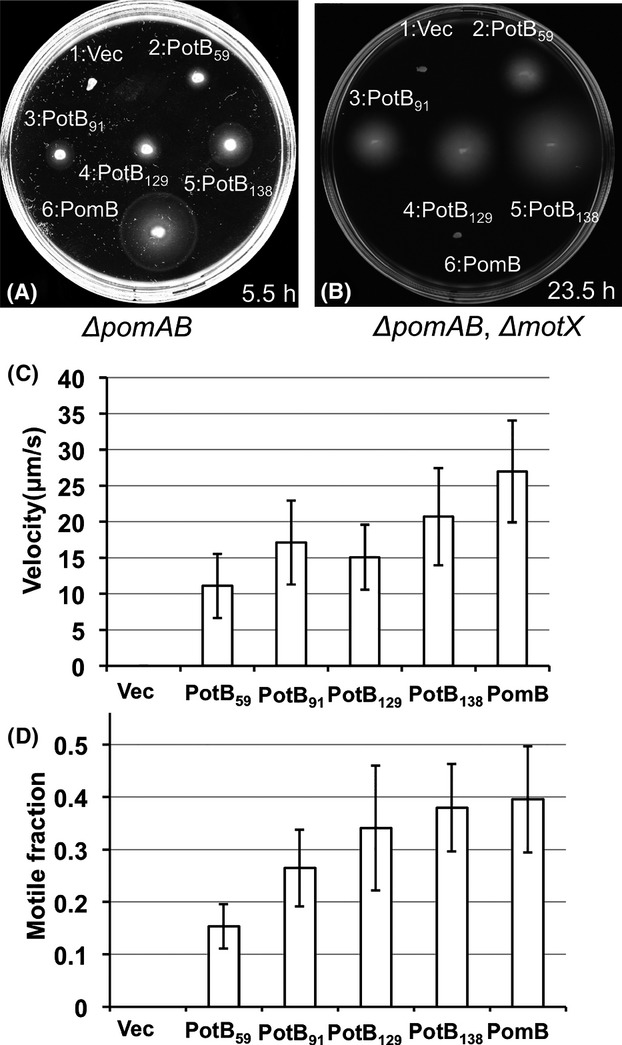
Motilities of chimeras in Vibrio. Fresh colonies of transformants from NMB191 (Vibrio alginolyticus ΔpomAB) (A) or TH4 (V. alginolyticus ΔpomAB ΔmotX) (B) were inoculated and incubated on 0.25% VPG agar for 5.5 h (A) and on 0.3% VPG agar for 23.5 h (B) at 30°C with 0.02% arabinose. The cells have plasmids: 1, pBAD33 (Vec); 2, pTF9 (PotB59); 3, pTSK62 (PotB91); 4, pTSK63 (PotB129); 5, pTSK64 (PotB138) and 6, pHFAB (PomB) and produce wild-type PomA and the PotB chimera or the PomB proteins. Swimming speeds (C) or motile fractions (D) of cells producing the wild-type PomA and PotB chimera or PomB proteins in NMB191 (V. alginolyticus ΔpomAB) were measured. After 4 h culture in VPG at 30°C with 0.02% (w/v) arabinose, the cells were observed under a microscope.
We next investigated the swimming ability in liquid medium (Fig.2C and D). PotB91, PotB129, and PotB59 in V. alginolyticus ΔpomAB conferred similar swimming speeds, about 15–20 μm/sec, and this speed was about half of the wild-type PomB. The swimming speed of PotB138 was faster than the other chimeric proteins and these swimming speeds correlated with the diameters of the swimming rings. The chimeric proteins in the V. alginolyticus ΔpomAB ΔmotX strain did not confer motility in most cells under the same conditions as ΔpomAB.
Expression profiles of the chimeric proteins in Vibrio
We examined the expression and stability of the chimeric proteins by immunoblotting using antibodies against MotB and PomA (Fig. S2). In regard to PomB or the chimeras, all chimeric proteins were detected but a band was not detected in the vector control or in wild-type PomB from the pHFAB plasmid in ΔpomAB ΔmotX. The PomB protein was not detected by the anti-MotB antibody. The band intensity of PotB91 was as high as that of PotB138, but the expression level of PotB129 was very low in both hosts. This may suggest that the PotB129 protein is unstable or is difficult to express. In contrast to the amounts of B subunits, the band intensities of PomA in the hybrid strains were similar in both hosts. The production of PomA was not affected by the chimeric potB genes.
Motility of E. coli by the chimeric proteins
We found that all strains produced chimeric proteins which conferred motility in the motX deficient strain, V. alginolyticus ΔpomAB ΔmotX. Thus, we next examined the influences of the swimming abilities if the chimeric proteins were expressed in E. coli ΔmotAB. The swimming abilities of chimera strains in soft agar plates were observed (Fig.3A and B). Only PotB59 showed swimming motility clearly in soft agar plates and the other chimera strains did not confer swimming rings even after 13 h of incubation in soft agar plates containing 0.25% agar. In regard to the swimming ability in liquid media, we examined the swimming speeds for all strains producing chimeric proteins (Fig.3C). PomB did not confer any motility even after overnight incubation in soft agar plates. Against our assumption, the swimming speeds of PotB91 and PotB138 were half as much as that of PotB59. This may suggest that the force-generating ability is not so different between PotB91 or PotB138 and PotB59. Next, because the swimming motilities of the new chimera strains on soft agar plates were different from that in liquid medium, we inferred that the reason for this difference might not be the swimming speed or torque. We thought that the most likely reason might be a defect in rotational switching, so that cells exhibited an imbalanced transition between smooth swimming – tumble states in the motility agar plates. When flagella, which are helical filaments, rotate counter-clockwise (CCW), cells show smooth swimming and when flagella rotate clockwise (CW), cells tumble. So, we examined the flagellar-rotational switching ability of the chimera strains using a tethered cell assay, in which we can detect rotational direction directly at the single motor level (Fig.3D). The switching ability of PotB59 was high since 60% of the cells could change directions in 30 sec. On the other hand, the switching abilities of PotB91 and PotB129 were less than 10% of PotB59 and that of PotB138 was not detectable in these conditions. All the chimeric motors rotated with a CCW bias. Therefore, we concluded that the difference between swimming on soft agar plates and in liquid medium of PotB91 and PotB138 strains was mainly caused by a deficiency of the switching property. We detected chimeric proteins in hybrid E. coli strains by immunoblotting (Fig. S3). The band intensities of the PotB91 and PotB138 bands were comparable to PotB59, but the intensity of the PotB129 band was much less than the others. Thus, the expression profiles of chimeric proteins in E. coli are similar to those in Vibrio.
Figure 3.
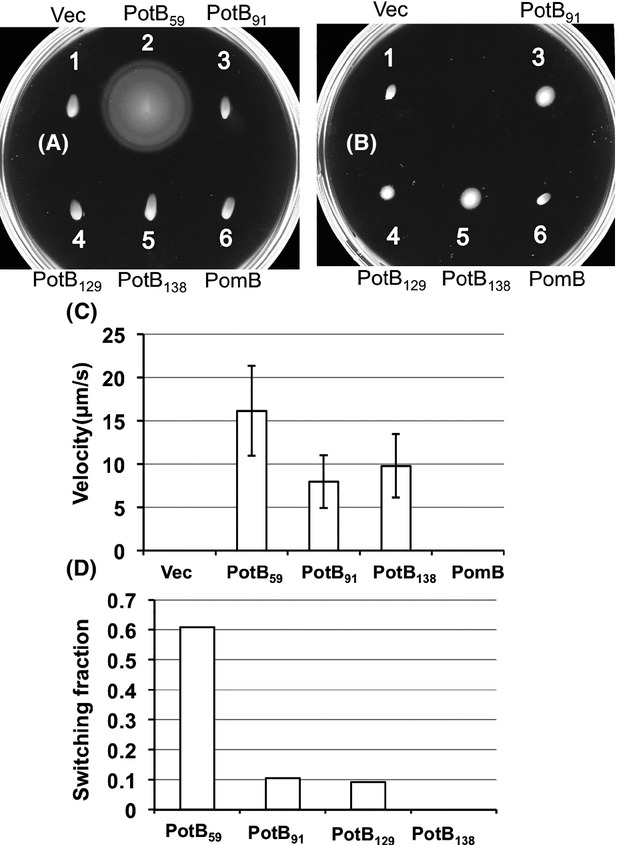
Motilities of chimeras in Escherichia coli. Fresh colonies of RP6894 (E. coli ΔmotAB) were inoculated and incubated on 0.3% TB agar for 13 h (A) and 20 h (B) at 30°C. Each strain has the following plasmid: 1, vector plasmid pBAD33 (Vec); 2, pTF9 (PotB59); 3, pTSK62 (PotB91); 4, pTSK63 (PotB129); 5, pTSK64 (PotB138); 6, pHFAB (PomB). (C) Swimming speeds of the chimera strains of RP6894 were measured. After 6 h of culture in TG medium with 0.02% (w/v) arabinose, the cells were diluted 15-fold with TG medium. Swimming speeds were measured as described in Experimental Procedures. (D) Rotational switching of the flagellar motor observed by the tethered cell assay. “Switching fraction” means the proportion of the cell number of rotating cells who changed the direction at least one time in 30 sec against the total number of rotating cells.
Suppressor mutants from the chimeras
Because of the results for E. coli ΔmotAB, we isolated spontaneous motile suppressors from the hybrid E. coli strains that were almost nonmotile in soft agar plates (Fig.4A). We isolated suppressors that showed significantly improved motility on soft agar plates compared to the original strain, and most of the suppressor mutations were mapped on the plasmid encoding both pomA and potB. We sequenced the plasmids to determine the mutation site (Fig.4B). For PotB138, suppressor 1 (sp1) carries M165I in PomA, sp2 has two mutations, A180T in PomA and E111K in PotB, and sp3 has D117N in PomA. For PotB91, sp1 has A57D in PotB. The diameter of PotB138-sp2 was the largest among the three suppressors of PotB138 and the diameter of PotB138-sp3 was the smallest among them. All mutations in PotB chimeras were mapped on the PomB region, so that the residue number is the PomB numeration.
Figure 4.
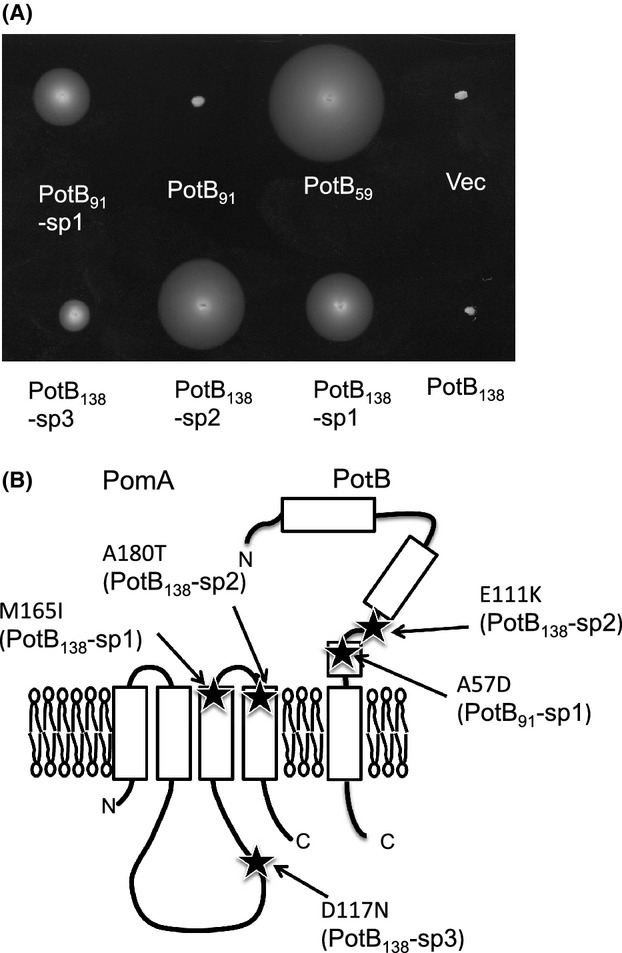
Suppressor mutants isolated from the chimera strains. (A) Swimming abilities in soft agar plates of chimeras in Escherichia coli were examined. Fresh colonies of RP6894 (E. coli ΔmotAB), which were retransformed with the plasmids recovered from the suppressor strains, were inoculated and incubated on 0.3% TB agar for 22–24 h at 30°C with 0.02% arabinose. 1, vector plasmid pBAD33 (Vec); 2, pTF9 (PotB59); 3, pTSK62 (PotB91); 4, PotB91-sp1; 5, pTSK64 (PotB138); 6, PotB138-sp1; 7, PotB138-sp2; 8, PotB138-sp3. (B) The mutation sites of the suppressors mapped on a topology model of PomA and PomB.
We examined the swimming speeds in liquid media of these suppressors (Fig.5). The swimming speed of PotB138-sp1 was slower than PotB138, but the swimming speeds of PotB138-sp2 and PotB138-sp3 were as fast as PotB138. The swimming speed of PotB91-sp1 was a little faster than PotB91. The swimming rings in soft agar plates were much increased but the swimming speeds of the suppressors were not changed. Thus, the suppressor mutations of PotB91 and PotB138 are suggested to affect the rotational switching of the flagellar motor mainly but not the force generation by the stator and rotor interaction.
Figure 5.
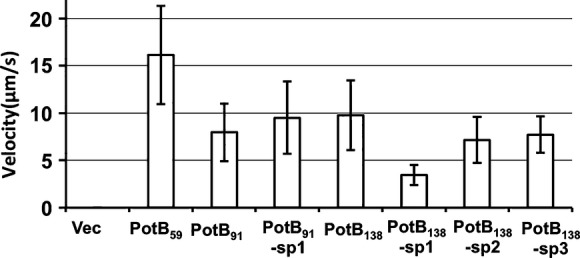
Swimming speeds of suppressors. After 4 h incubation in TG medium with 0.02% (w/v) arabinose, the cells were observed under a microscope. For details of this analysis, see Experimental Procedures.
Discussion
It was reported that the ΔpomAB mutant (V. alginolyticus NMB191), which carries a plasmid (pHFAB) containing the pomA and pomB genes, can swim well but that the ΔpomAB ΔmotX mutant (V. alginolyticus TH4), which carries the plasmid pHFAB, cannot swim at all (Asai et al. 2003; Terashima et al. 2006). When MotX or MotY is missing, the stators composed of PomA and PomB cannot assemble around the flagellar rotor in Vibrio. A functional chimera PotB (renamed in this study as PotB59) with PomA conferred motility to either the ΔpomAB ΔmotX mutant or the ΔpomAB mutant (Asai et al. 2003). The new chimeric proteins of PotB91, PotB129 and PotB138 conferred motility to either the ΔpomAB ΔmotX mutant or the ΔpomAB mutant (Fig.2). The motility without MotX in soft agar plates is severely reduced and we could not detect swimming cells that were deficient MotX in liquid medium until 4 h after the overnight culture was inoculated into fresh broth at a 100-fold dilution. Without the T ring, the swimming fraction of PotB chimera strains is very small, and it is much smaller than that with the T ring. We assume that the PotB chimeric proteins of the stator can assemble around the rotor but the basal body without the T ring probably is not able to rotate smoothly. We cannot rule out that the PotB chimeric proteins are not efficiently assembled or installed into the motor in the T ring deficient cells.
Although the expression level of PotB129 was very low in the chimeric proteins (Fig. S2), PotB129 conferred swimming ability both in soft agar plates and in liquid media similar to PotB91. The swimming speed of PotB138 was comparable to that of the wild-type PomB. Although we cannot rule out that the difference in motility among chimera strains is mainly due to the low expression of PotB129, the expression of PotB129 seems to be sufficient to make adequate stators since the induction by arabinose 0.02% is high and the protein level is higher than the chromosomal expression level. Among the chimeric proteins, PotB138 conferred better motility than the others in the ΔpomAB mutant as well as in the ΔpomAB ΔmotX mutant. The α1 and α2 helices of PotB138 are derived from Vibrio PotB138. This may suggest that the α1 and α2 helices are important for motility in Vibrio and we speculate that the helices or the region around the helices interact with MotX. We had speculated previously that the structure of an N-terminal region of PomBC (the N terminal PEM region of PomB, 121–153) was disordered when the PomAB stator complex was activated (Zhu et al. 2014). The exact role of the helices remains unclear and it is possible that the helices interact with a flagellar protein other than MotX.
We have shown that PotB59 with PomA conferred motility on ΔmotAB E. coli and that E. coli strains are driven by sodium ions and not by protons (Asai et al. 2003). The swimming motility of PotB chimera strains in Vibrio in soft agar plates was smaller than that of PomB. However, the swimming motility of PotB in liquid medium seems not to be different from that of PomB. The swimming speed in liquid medium is almost linearly proportional to the ratio of the swimming fraction except for PotB129. The protein expression level of PotB129 was very low compared to the other chimeras in both E. coli and V. alginolyticus (Fig. S2A). In contrast to Vibrio, E. coli with the new chimera PotB did not show swimming ability in soft agar plates but could swim in liquid medium. The tethered cell assay showed that their switching abilities were much worse than PotB59 (Fig.3D). The B subunit of the stator has a linker region from the region posterior to the plug to the region prior to the α1 helix. The linker regions of PotB91 or PotB138 are derived from PomB and the linker region of PotB59 is derived from MotB. The linker region may affect the switching between CCW and CW rotation which is believed to be caused by a conformational change of the FliG C-terminal region (Lloyd et al. 1999; Lee et al. 2010; Minamino et al. 2011). It is inferred that a specific interaction between the FliG C-terminal region and the cytoplasmic loop region of MotA or PomA determines the direction of the force generation (Zhou et al. 1998a; Yorimitsu et al. 2002; Paul et al. 2011; Takekawa et al. 2014). We have shown that some mutations of the charged residues of PomA and FliG cause a defect in the rotational switching of the motor (Takekawa et al. 2014). The suppressor mutations were isolated from PotB91 or PotB138 in E. coli. For PotB138, the mutations are M165I, D117N and A180T in PomA or E111K in PotB. For PotB91, the mutation is A57D in PotB. The mutations are located in the plug or the linker region of the PotB proteins, in the periplasmic boundary region of PomA TM3 or TM4, and in a cytoplasmic region of PomA. Because MotBC is required not only for proper anchoring of the stator to its binding site on the motor (Blair et al. 1991) but also for proper alignment of the stator relative to the rotor (Garza et al. 1996), these genetic data suggest the possibility that the suppressor mutations may affect the stator–rotor interface of the PomA/PotB chimeric motor in E. coli, allowing the motor to spin in both the CCW and CW directions. We showed that MotB allows the E. coli MotA/B complex to efficiently function as a H+-coupled stator in Vibrio and hyper-motile suppressions were isolated in the periplasmic region of MotB (Asai et al. 2003). The mechanism was not assigned in that study, but the suppression may similarly occur as the suppression of this study.
Acknowledgments
We thank Hajime Fukuoka for providing the plasmid. We thank Akiko Abe and Shiwei Zhu for technical assistance. This work was supported by the Japan Society for the Promotion of Science (JSPS) of KAKENHI Grant Numbers 24117004 and 23247024 to M. H., KAKENHI Grant Number 24657087 to S. K. Y. N. was partly supported by the Integrative Graduate Education and Research program of Nagoya University.
Conflict of Interest
None declared.
Supporting Information
Sequence alignment based on the secondary structures of chimeric proteins refer to the crystal structures of PomBC5 and MotBC2 (PDB ID codes: 3WPW and 2ZVY). They show junction sites of chimera strains and important motifs or domains. Blue underlines show α helices in regions whose crystal structures are solved and green underlines show β sheets. The black box shows the TM region and the purple box shows the plug region.
Figure S2. Immunoblotting of chimeric proteins expressed in Vibrio. Whole cells of NMB191 (Vibrio alginolyticus ΔpomAB) and TH4 (V. alginolyticus ΔpomAB ΔmotX) containing the vector plasmids pBAD33 (Vec), pTF9 (PotB59), pTSK62 (PotB91), pTSK63 (PotB129), pTSK64 (PotB138) or pHFAB (PomB) were subjected to SDS-PAGE, followed by immunoblotting using anti-Salmonella MotBC (A) and anti-PomA antibodies (B).
Figure S3. Immunoblotting of chimera proteins expressed in Escherichia coli. Whole cells of E. coli ΔmotAB (RP6894) containing the vector plasmid pBAD33 (Vec), pTF9 (PotB59), pTSK62 (PotB91), pTSK63 (PotB129), pTSK64 (PotB138) or pHFAB (PomB) were subjected to SDS-PAGE and immunoblotting was performed using an anti-Salmonella MotB antibody.
References
- Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I. Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai Y, Yakushi T, Kawagishi I. Homma M. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 2003;327:453–463. doi: 10.1016/s0022-2836(03)00096-2. [DOI] [PubMed] [Google Scholar]
- Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545:86–95. doi: 10.1016/s0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- Blair DF, Kim DY. Berg HC. Mutant MotB proteins in Escherichia coli. J. Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun SY. Parkinson JS. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Fukuoka H, Yakushi T, Kusumoto A. Homma M. Assembly of motor proteins, PomA and PomB, in the Na+-driven stator of the flagellar motor. J. Mol. Biol. 2005;351:707–717. doi: 10.1016/j.jmb.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Garza AG, Biran R, Wohlschlegel JA. Manson MD. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ. Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking ER, Vogt C, Bakker EP. Manson MD. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 2006;364:921–937. doi: 10.1016/j.jmb.2006.09.035. [DOI] [PubMed] [Google Scholar]
- Kojima S. Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- Kojima S, Shinohara A, Terashima H, Yakushi T, Sakuma M, Homma M, et al. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. USA. 2008;105:7696–7701. doi: 10.1073/pnas.0800308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, et al. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009;73:710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- Lee LK, Ginsburg MA, Crovace C, Donohoe M. Stock D. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature. 2010;466:996–1000. doi: 10.1038/nature09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Kojima S. Homma M. Characterization of the periplasmic region of PomB, a Na+-driven flagellar stator protein in Vibrio alginolyticus. J. Bacteriol. 2011a;193:3773–3784. doi: 10.1128/JB.00113-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Kojima S. Homma M. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells. 2011b;16:985–999. doi: 10.1111/j.1365-2443.2011.01545.x. [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Whitby FG, Blair DF. Hill CP. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- Minamino T, Imada K, Kinoshita M, Nakamura S, Morimoto YV. Namba K. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 2011;9:e1000616. doi: 10.1371/journal.pbio.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto YV. Minamino T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules. 2014;4:217–234. doi: 10.3390/biom4010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto YV, Nakamura S, Kami-ike N, Namba K. Minamino T. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 2010;78:1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- Morimoto YV, Nakamura S, Hiraoka KD, Namba K. Minamino T. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J. Bacteriol. 2013;195:474–481. doi: 10.1128/JB.01971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M. Kojima S. Bacterial motility measured by a miniature chamber for high-pressure microscopy. Int. J. Mol. Sci. 2012;13:9225–9239. doi: 10.3390/ijms13079225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Yakushi T. Homma M. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J. Biol. Chem. 2005;280:25659–25664. doi: 10.1074/jbc.M500263200. [DOI] [PubMed] [Google Scholar]
- Paul K, Brunstetter D, Titen S. Blair DF. A molecular mechanism of direction switching in the flagellar motor of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2011;108:17171–17176. doi: 10.1073/pnas.1110111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. USA. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. Homma M. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 2000;275:20223–20228. doi: 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- Sudo Y, Kitade Y, Furutani Y, Kojima M, Kojima S, Homma M, et al. Interaction between Na+ ion and carboxylates of the PomA-PomB stator unit studied by ATR-FTIR spectroscopy. Biochemistry. 2009;48:11699–11705. doi: 10.1021/bi901517n. [DOI] [PubMed] [Google Scholar]
- Takekawa N, Terauchi T, Morimoto YV, Minamino T, Lo CJ, Kojima S, et al. Na+ conductivity of the Na+-driven flagellar motor complex composed of unplugged wild-type or mutant PomB with PomA. J. Biochem. 2013;153:441–451. doi: 10.1093/jb/mvt011. [DOI] [PubMed] [Google Scholar]
- Takekawa N, Kojima S. Homma M. Contribution of many charged residues at the stator-rotor interface of the Na+-driven flagellar motor to torque generation in Vibrio alginolyticus. J. Bacteriol. 2014;196:1377–1385. doi: 10.1128/JB.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Fukuoka H, Yakushi T, Kojima S. Homma M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol. Microbiol. 2006;62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- Terashima H, Koike M, Kojima S. Homma M. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J. Bacteriol. 2010a;192:5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kojima S. Homma M. Functional transfer of an essential aspartate for the ion-binding site in the stator proteins of the bacterial flagellar motor. J. Mol. Biol. 2010b;397:689–696. doi: 10.1016/j.jmb.2010.01.050. [DOI] [PubMed] [Google Scholar]
- Terauchi T, Terashima H, Kojima S. Homma M. A conserved residue, PomB-F22, in the transmembrane segment of the flagellar stator complex, has a critical role in conducting ions and generating torque. Microbiology. 2011;157:2422–2432. doi: 10.1099/mic.0.048488-0. [DOI] [PubMed] [Google Scholar]
- Yakushi T, Yang J, Fukuoka H, Homma M. Blair DF. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 2006;188:1466–1472. doi: 10.1128/JB.188.4.1466-1472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T. Homma M. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Sato K, Asai Y, Kawagishi I. Homma M. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 1999;181:5103–5106. doi: 10.1128/jb.181.16.5103-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Sowa Y, Ishijima A, Yakushi T. Homma M. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 2002;320:403–413. doi: 10.1016/S0022-2836(02)00426-6. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lloyd SA. Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 1998a;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, et al. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 1998b;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Takao M, Li N, Sakuma M, Nishino Y, Homma M, et al. Conformational change in the periplasmic region of the stator coupled with the assembly around the rotor. Proc. Natl. Acad. Sci. USA. 2014;111:13523–13528. doi: 10.1073/pnas.1324201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment based on the secondary structures of chimeric proteins refer to the crystal structures of PomBC5 and MotBC2 (PDB ID codes: 3WPW and 2ZVY). They show junction sites of chimera strains and important motifs or domains. Blue underlines show α helices in regions whose crystal structures are solved and green underlines show β sheets. The black box shows the TM region and the purple box shows the plug region.
Figure S2. Immunoblotting of chimeric proteins expressed in Vibrio. Whole cells of NMB191 (Vibrio alginolyticus ΔpomAB) and TH4 (V. alginolyticus ΔpomAB ΔmotX) containing the vector plasmids pBAD33 (Vec), pTF9 (PotB59), pTSK62 (PotB91), pTSK63 (PotB129), pTSK64 (PotB138) or pHFAB (PomB) were subjected to SDS-PAGE, followed by immunoblotting using anti-Salmonella MotBC (A) and anti-PomA antibodies (B).
Figure S3. Immunoblotting of chimera proteins expressed in Escherichia coli. Whole cells of E. coli ΔmotAB (RP6894) containing the vector plasmid pBAD33 (Vec), pTF9 (PotB59), pTSK62 (PotB91), pTSK63 (PotB129), pTSK64 (PotB138) or pHFAB (PomB) were subjected to SDS-PAGE and immunoblotting was performed using an anti-Salmonella MotB antibody.


