Fig. 7. 13C-methyl thiocystine (MTC) ClC-ec1.
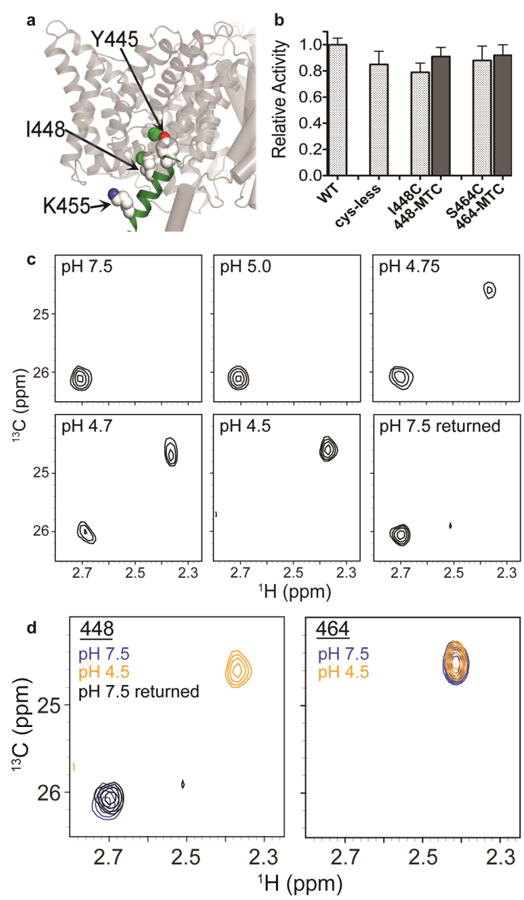
a Position of helix R and I448. The position of helix R in one subunit of the ClC-ec1 homodimer is shown in green. The intracellular Cl- gate residue Y445 (shown in spacefill) is the first residue of Helix R. 13C-MTC probes were introduced at positions I448 (shown in spacefill) and S464 (not shown; part of the unresolved C-terminus of ClC-ec1). 13C-K455 (spacefilled) was investigated by 13C-Lys methylation (Figure 4).
b Activity assays on 13C-MTC ClC-ec1 variants, with Cl- flux assays were performed as in Figure 6. Activity is normalized to WT ClC-ec1. Cys mutants were introduced into a cys-less template ClC-ec1 (Nguitragool and Miller 2007). Labeling at I448C and at S464C (with labeling efficiencies 93±5% and 89±3% respectively) has no effect on activity. Data represent averages ± SEM for n=8-11 replicates.
c1H-13C HSQC spectra of 13C-I448MTC ClC-ec1 at pH 7.5, 5.0, 4.75, 4.7, 4.5, and reversed to pH 7.5. At pH 7.5 and pH 5.0, the 13C-MTC probe exhibits a single peak (δH = 2.70 ppm). At pH 4.75 and pH 4.7, a second peak at δH = 2.36 ppm is additionally observed, and at pH 4.5 only this second peak remains. On reversal to pH 7.5, the spectrum reverts to the single peak at δH = 2.70 ppm.
d1H-13C HSQC spectra of 13C-I448MTC ClC-ec1 (left) and 13C-S464MTC (right) at pH 7.5 (blue), pH 4.5 (orange) and (for I448-MTC) reversed to pH 7.5 (black). S464 resides in the unresolved C-terminus of the protein. Its insensitivity to pH changes suggests that the peak shift in observed with I448 is likely due to structural changes in the protein rather than a general effect of pH.
