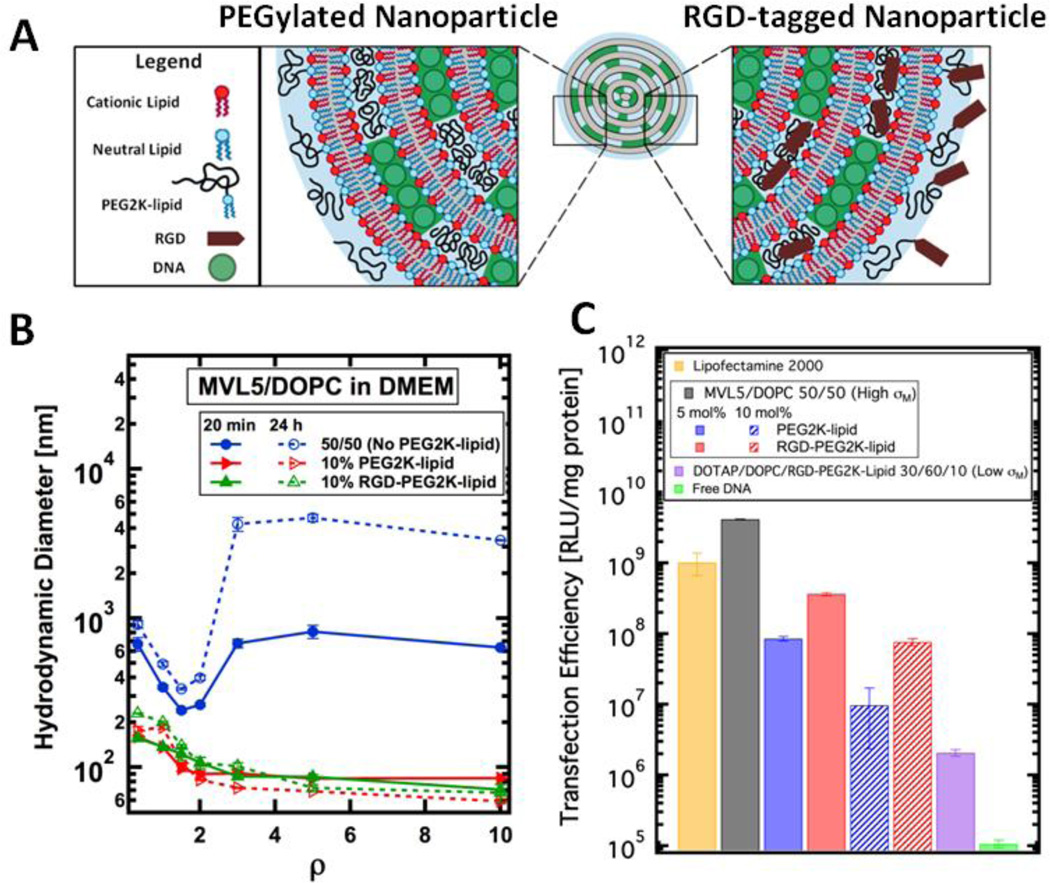
Figure 2.
Size and transfection efficiency (TE) of MVL5-based CL–DNA complexes and NPs. (A) Schematic showing the internal nano-structure and functionalized surface of a PEGylated and RGD-tagged CL–DNA nanoparticle. (B) Dynamic light scattering shows that CL–DNA complexes lacking PEGylation form micron-sized aggregates when complexed in the cell culture medium DMEM. (blue curve). PEGylation with 10 mol% PEG2K-lipid or RGD-PEG2K-lipid induces the formation of sub-200 nm, sterically-stabilized nanoparticles (red and green curves). (C) MVL5/DOPC complexes show remarkably high TE, outperforming the commercial reagent Lipofectamine® 2000. As the concentration of PEG2K-lipid increases (blue bars), TE decreases. RGD-tagging of MVL5-based NPs partially recovers TE (red bars) relative to the PEGylated NPs lacking RGD. Low-σM NPs contain 30/60/10 DOTAP/DOPC/RGD-PEG2Klipid by mol% and show low TE.