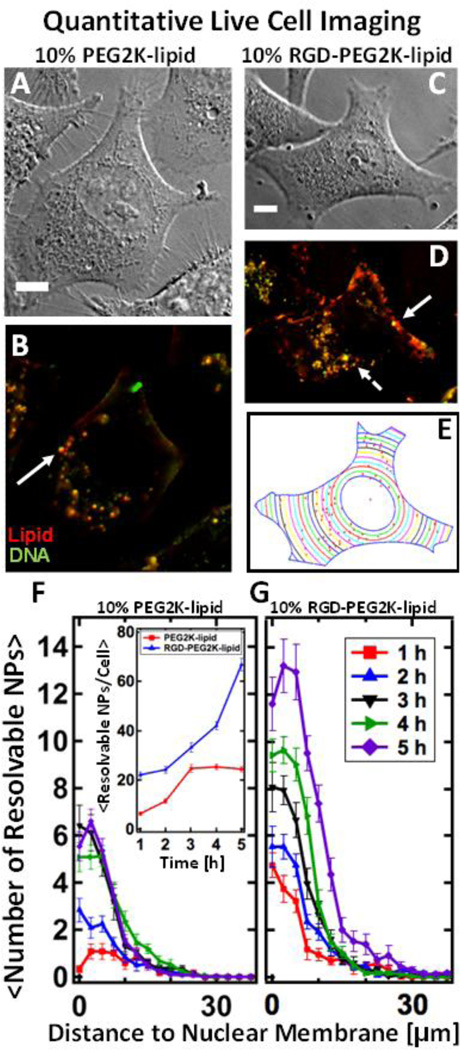
Figure 3.
Live-cell imaging of fluorescently-labeled PEGylated MVL5-based nanoparticles with and without RGD-tagging. NPs containing multivalent lipid (MVL5/DOPC/(PEG2K-lipid or RGD-PEG2K-lipid) at 50/40/10 mol% at ρ=10) are internalized in live L-cells. (Red – TRITC-DHPE (lipid), Green – Cy5 (DNA), Yellow – Nanoparticle composed of complexed lipid and DNA) (A, B) DIC and fluorescent images of Lcells after 4 hours of incubation with PEGylated NPs which lack RGD. (C, D) DIC and fluorescent images of L-cells after 4 hours of incubation with RGD-tagged NPs. Relative to NPs lacking RGD (A, B), RGDtagging causes a higher number of NPs to bind to the plasma membrane as well as internalize. Large, bright fluorescent particles (solid arrows) are likely endosomes containing multiple NPs which are spatially irresolvable. The dashed arrow points to a tiny resolution-limited spot which is believed to be a single NP. (E) A MatLab routine was used to determine the boundary and nuclear location of the cell in (C). Using (D), the routine locates all intracellular particles (red spots in (E)) and measures their distance to the nucleus. (F, G) The results of the localization routine for NPs without and with RGD respectively. By defining the nuclear membrane as a reference point, we can average the localization results over 20 cells. Spatial localization shows that while both types of MVL5 NPs (with and without RGD-tagging) are uptaken and trafficked to the perinuclear region, approximately three times as many RGD-tagged particles are found in cells after 5 hours of incubation (a standard incubation time for a TE assay). Inset shows total NPs per cell. All scale bars are 10 µm.