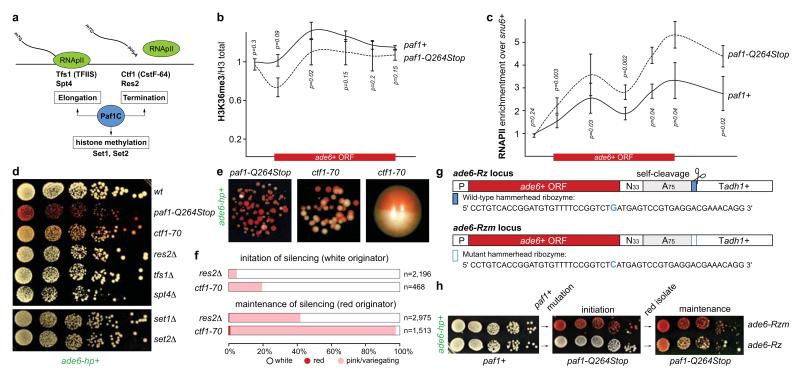
Figure 3. Mechanism of repression.
a, Paf1C governs RNA Pol II transcription elongation, RNA 3’-end processing, and transcription-coupled histone modifications. b and c, ChIP experiments to assess ade6+ transcriptional activity in Paf1C mutant cells. H3K36me3 levels were normalized to total H3 levels. snu6+ is transcribed by RNAPIII and serves as background control. Error bars, SEM; n=3 independent biological replicates; p values were calculated using one-tailed Student’s t test. d and e, Silencing assays showing that ade6+ siRNAs can initiate repression of ade6+ in transcription termination mutants. Note the bi-stable state of repression in ctf1-70 cells. Cells were grown on yeast extract (YE) plates. f, Percentage of naïve transcription termination defective cells (white originator) that establish heterochromatin within 20-30 mitotic divisions (initiation) and stability of ectopic heterochromatin in descendants thereof (maintenance). n, number of scored colonies. g, A 52-mer wild-type or mutant hammerhead ribozyme sequence preceded by a templated polyA(75)-tail was integrated 33nt downstream of the ade6+ stop codon (ade6-Rz or ade6-Rzm, respectively). h, Silencing assay showing that ade6+ siRNAs stably repress ade6-Rzm but not ade6-Rz in paf1 mutant cells. Note that ade6-Rz produces fully functional mRNA in paf1+ cells.