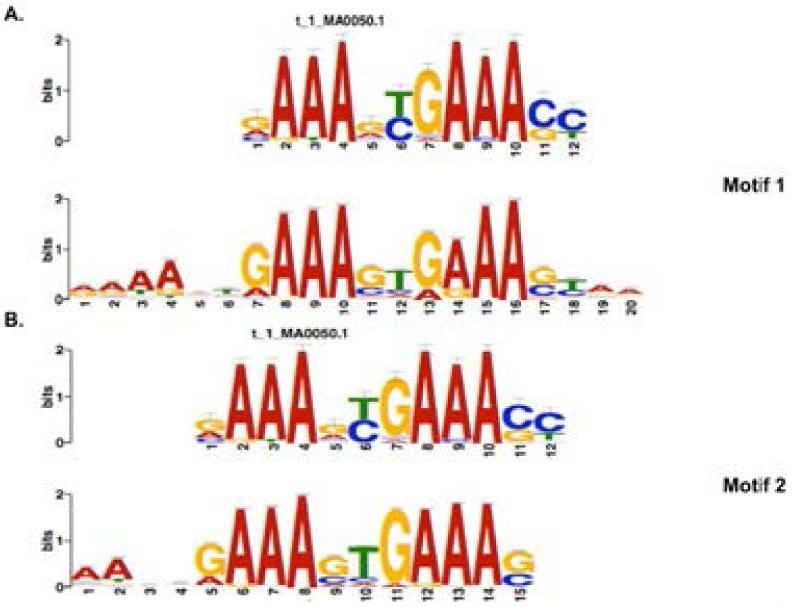
Figure 5.
De novo motif discovery carried out on IFN-gamma stimulated peak regions reveals new determinants for IRF1 binding A. Shown is the logo for Motif 1 which is the most statistically significant DNA motif derived from all peak regions found in IFN-gamma stimulated cells (e-value, 1.2e-168) B. Shown is the logo for Motif 2 which is the most statistically significant DNA motif derived from IFN-gamma unique peak regions (only found in IFN-gamma stimulated cells)(e-value, 7.0e-455). Both motifs are aligned to the consensus IRF1 motif (MEME-ChIP).