Abstract
The proteolytic system of Lactobacillus bulgaricus breaks down milk proteins into peptides and amino acids, which are essential for the growth of the bacteria. The aim of this study was to determine the expressions of seven key genes in the proteolytic system under different culturing conditions (different phases, initial pH values, temperatures, and nitrogen sources) using real-time polymerase chain reaction (RT-PCR). The transcriptions of the seven genes were reduced by 30-fold on average in the stationary phase compared with the exponential growth phase. The transcriptions of the seven genes were reduced by 62.5-, 15.0-, and 59.0-fold in the strains KLDS 08006, KLDS 08007, and KLDS 08012, respectively, indicating that the expressions of the seven genes were significantly different among strains. In addition, the expressions of the seven genes were repressed in the MRS medium containing casein peptone. The effect of peptone supply on PepX transcription was the weakest compared with the other six genes, and the impact on OppD transcription was the strongest. Moreover, the expressions of the seven genes were significantly different among different strains (P<0.05). All these results indicated that the culturing conditions affected the expression of the proteolytic system genes in Lactobacillus bulgaricus at the transcription level.
Keywords: Gene expression, Proteolytic system, Lactobacillus bulgaricus, Proteinase
1. Introduction
Lactic acid bacteria (LAB) have been used for centuries as a starter in dairy fermentations (Chen et al., 2014). The degradation of milk proteins through proteolysis by LAB plays an important role in generating peptides and amino acids for bacterial growth (Kunji et al., 1996; Savijoki et al., 2006). The proteolytic system of LAB consists of extracellular peptidases, peptide transport system, and intracellular peptidases. (1) Proteolysis of casein (milk proteins) by LAB is initiated by a cell-wall bound proteinase (Prt), which degrades the extracellular casein into peptides (Roland, 1999; Chen et al., 2003). The only cell-wall bound proteinase in Lactobacillus delbrueckii subsp. bulgaricus is PrtB (Gilbert et al., 1996; Stefanitsi and Garel, 1997). (2) The peptides are subsequently transported by several peptide transport systems into cells; oligopeptides are transported by the oligopeptide transport system (Opp) and the di/tri-peptides are transported by a proton motive force-driven transporter DtpT or an adenosine triphosphate (ATP)-driven Dpp system (Doeven et al., 2005; Picon et al., 2010). In Lactobacillus delbrueckii subsp. bulgaricus, the peptide transport system is Opp and Dpp. (3) After peptides are taken up into the cells, they are degraded into amino acids by concerted actions of intracellular peptidases including aminopeptidases that cleave amino acids from the N-terminus of the peptides, endopeptidases that cleave the internal peptide bonds, and di- and tri-peptidases that degrade di- and tri-peptides, respectively (Liu et al., 2010). In particular, caseins are rich in proline and LAB have numerous proline peptidases for degrading proline-rich peptides (Stucky et al., 1995; Christensen et al., 1999).
Lactobacillus bulgaricus can decrease bitterness and produce characteristic flavor to acidified milk, which makes this bacterium an important component of starter cultures for the dairy industry (Griffiths and Tellez, 2013). Milk proteins are the most primary source of bioactive peptides that can be produced through hydrolysis by Lactobacillus helveticus. As a result of proteolysis, fermented milk has a higher content of peptides and free amino acids, especially histidine, valine, proline, and serine, than non-fermented milk (Matar et al., 2003). LAB obtain all the necessary free amino acids required for growth in milk by their complicated proteolytic system. In a more recent study, the expressions of six transcriptional units, including PepD, PepN, PepC, PepX, PrtP, and Opp-PepO1, were found to be repressed by 5- to 150-fold with the addition of casein hydrolysate containing 20% amino acids and 80% peptides to the growth medium and the expression was released only when cells encountered nitrogen-limiting conditions (Guédon et al., 2001). Environmental factors are also known to influence the expression of certain peptidase genes. For example, the PepR expression of Lactobacillus sakei was repressed during aerobic growth but induced 20-fold under anaerobic conditions (Champomier-Vergès et al., 2002).
Because of the important role of the proteolytic system of LAB, numerous studies have been carried out to characterize the proteins and determine the factors that influence their expression. The operon of the putative (Opp) of Lactobacillus delbrueckii subsp. bulgaricus B14 was cloned and characterized (Peltoniemi et al., 2002). Other peptidases and transport proteins of the proteolytic system were also characterized (Mayo et al., 1991; Tynkkynen et al., 1993; Kunji et al., 1995; Savijoki and Palva, 2000). Expressions of PrtP (proteinase precursor), PrtM (maturase), several peptidases and transport proteins of the proteolytic system during the growth in skim milk were investigated (Azcarate-Peril et al., 2009; Wu et al., 2011). The gene expression dynamics of the PrtP and PrtM of Lactobacillus casei Zhang during fermentation in soymilk was also studied, and real-time polymerase chain reaction (PCR) was used to analyze the differentially expressed genes at the lag phase, the late logarithmic phase, and the stationary phase (Wang et al., 2012). Furthermore, the expressions of Opp, DtpT, and PepT in the presence of different peptides were investigated, and it was found that the growth phase and the peptide source significantly affected the gene transcription (Guédon et al., 2001; Vermeulen et al., 2005).
In this study, we used the Lactobacillus bulgaricus to explore the expressions of seven genes in the proteolytic system at different growth phases, initial pH values, culturing temperatures, and nitrogen sources by real-time PCR. Our research suggested that the gene expressions were significantly different under different conditions. These results are valuable for the effective use of the strains.
2. Materials and methods
2.1. Strains and growth conditions
The Lactobacillus bulgaricus strains used in this study are listed in Table 1. They were from the Key Laboratory of Dairy Science, Ministry of Education, Northeast Agricultural University, Harbin, China. The strains were previously isolated from naturally fermented milk and koumiss made by herdsman families in the Tibet Autonomous Region, the Xinjiang Uygur Autonomous Region, and Gansu and Qinghai Provinces of China. The strains that had high growth and glucose catabolism rates showed serious post acidification, and the strains that had lower tolerance to acid showed weaker post acidification. The different Lactobacillus bulgaricus strains were available as frozen (−80 °C) stock cultures. They were revived by growth at 42 °C in normal deMan, Rogosa, and Sharpe medium (MRS; composition per liter: 20 g glucose·H2O, 10 g peptone, 10 g beef extract, 5 g yeast extract, 2.0 g KH2PO4·3H2O, 5.0 g CH3COONa, 2.0 g (NH4)2HC6H5O7, 0.58 g MgSO4·7H2O, 0.25 g MnSO4·H2O, 1 ml Tween 80, and 15 g agar for solid media). The revived strains were stored at 4 °C until required for experiments. The growth and proteolytic activity of LAB are affected by some factors, for example, strains species, type and concentration of cultivation nitrogen, time, temperature, and initial pH. So the conditions were selected to determine the impact on key enzyme gene expression. In the MRS medium containing extra peptone, 10 g/L casein peptone was added. The initial pH of MRS was adjusted to 5.6 and 6.5, and the culture temperature was set up to 30, 37, 42, and 45 °C, respectively, for culturing strains in the experiment.
Table 1.
Lactobacillus bulgaricus strains used in this study
| No. | Strain name | Strain No. |
| 1 | Lactobacillus delbrueckii subsp. bulgaricus | KLDS 08006 (1.0207)* |
| 2 | Lactobacillus delbrueckii subsp. bulgaricus | KLDS 08007 (1.0501) |
| 3 | Lactobacillus delbrueckii subsp. bulgaricus | KLDS 08012 (1.1007) |
The number of the strain is the preservation number of Dairy Industry Culture Collection of Key Laboratory of Dairy Science, Ministry of Education, Northeast Agricultural University, Harbin, China
2.2. Proteolytic activity in reconstituted skim milk (RSM)
Proteolytic activity by LAB was measured every 2 h from 0 to 12 h by the o-phthaldialdehyde (OPA) test (Church et al., 1983). The increase in optical density at 340 nm (OD340) relative to the control was determined using an ultraviolet spectrophotometer (UV1601, Shimadzu, Japan). All the bacterial strains were grown overnight at 37 °C and pH 6.2 (OD=0.70). Cells (2%, v/v) were inoculated into 12% (0.12 g/ml) RSM and incubated at their optimum culture condition. The 2-ml sample of each incubated RSM was mixed with 4 ml of 0.75 mol/L trichloroacetic acid (TCA). The cells were collected from the growth medium by centrifugation at 8000g for 10 min at 4 °C. The supernatant aliquot of this mixture was added to 3 ml of OPA reagent and incubated at room temperature for 5 min until the OD340 was read in the spectrophotometer, and 3 ml aliquots of the samples at each time point were selected.
2.3. Total RNA isolation and cDNA synthesis
The fermented samples, cut into small pieces, were flash frozen in liquid nitrogen to inhibit further transcription and stored at −80 °C until use. Just before isolation, samples were thawed at 4 °C. At no stage was the temperature of the samples allowed to exceed 4 °C. The fermented samples (10 g) were homogenized in 90 ml of 2% (0.02 g/ml) trisodium citrate solution at 4 °C. Cells from 10 ml of suspension were then recovered by centrifugation for 5 min at 6000g and at 4 °C (Ulve et al., 2008; Genay et al., 2009).
RNA was isolated from each sample using the RNeasy Midi Kit (TIANGEN Biotech (Beijing) Co., Ltd., China) according to the manufacturer’s instructions. The RNA templates were eluted in 40 ml RNase-free water. The quality and integrity of the total RNA templates were assessed by electrophoresis on 2% (0.02 g/ml) denaturing formaldehyde agarose gel. Complementary DNA (cDNA) was synthesized using the cDNA RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. The reverse transcriptase reaction mixture consisted of 8 μl PrimeScript® Buffer, 2 μl PrimeScript® RT Enzyme Mix Ι, 2 μl Oligo dT Primer (50 μmol/L), 2 μl Random 6 mers (100 μmol/L), 20 μl Total RNA, and 6 μl RNase Free dH2O. The total volume was 40 μl. The reverse transcriptase reaction conditions were as follows: 37 °C for 15 min and 85 °C for 5 s.
2.4. Real-time PCR
Primers of corresponding genes as shown in Table 2 were designed based on the published genome sequences of Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 (GenBank accession number: NC_008054.1) and gene quantification was performed with 16S ribosomal RNA (16S rRNA) as an internal standard. Real-time PCR was performed in a LightCycler instrument (ABI PRISM® 7500 Real-Time PCR System, Applied Biosystems, USA) according to the manufacturer’s instructions. Samples were subjected to 30 s denaturation at 95 °C, followed by 40 cycles of 5 s at 95 °C and 34 s at 60 °C. The reaction mixture consisted of 10.0 μl SYBR® Premix Ex Taq™ (Perfect Real Time; 2×Conc.; TaKaRa, Dalian, China), 2.0 μl cDNA, 6.8 μl dH2O, 0.4 μl ROX Reference Dye II (50×), and 0.4 μl of each primer (final concentration, 10 μmol/L). The total volume was 20 μl. The data were analyzed using the ABI PRISM 7500 System Sequence Detection software. The relative gene expression was calculated from three independent experiments and analyzed using the 2−∆∆CT value method (Livak and Schmittgen, 2001; Niesters, 2001; Schmittgen and Livak, 2008). An application plot and dissociation curves were used for the examination of the amplified products.
Table 2.
Oligonucleotide pairs used for determining gene expression of Lactobacillus bulgaricus
| Protein | Gene | Primer sequence (5' to 3') | Product (bp) | T m (°C) |
| 16S ribosomal RNA | 16S rRNA | Sense: ATCGGAAACTGTCATTCTTG; Antisense: CTAATCCTGTTCGCTACCC | 157 | 51.6 |
| Oligopeptide transporter | OppD | Sense: CCAGTTTGAACCCTTTGA; Antisense: CTTAGGCATCCCTACTTGAT | 128 | 51.3 |
| Cell-envelope proteinase B | PrtB | Sense: GTGGCAATTCAAATCTAATC; Antisense: GCTGTAATCATCACCCTCAT | 107 | 47.9 |
| Aminopeptidase C | PepC | Sense: CAAGCCGACTAAGTGGAAG; Antisense: AACAGCCTGGTAAACGAA | 109 | 50.1 |
| Endopeptidase F | PepF | Sense: TATCGCAGTGAGGTTCC; Antisense: TTCTTCTGGTTGCTGGTGT | 254 | 53.8 |
| Proline peptidase Q | PepQ | Sense: ACTGATAAGATGCGGGAAAT; Antisense: GCAAAGCCTGGGATGTAG | 275 | 54.8 |
| Proline peptidase X | PepX | Sense: TATTCAACGGCAAGTCCC; Antisense: TCCCGCCAAAGTAAGGTG | 195 | 54.9 |
| Tripeptidase T | PepT | Sense: TACGGACGGGTCACAACT; Antisense: GTCAACAGCCTTCCACATT | 121 | 51.7 |
T m: melting temperature
2.5. Statistical analysis
Data were expressed as the mean±standard deviation (SD) of triplicates. The statistical significance of the difference between two means was evaluated using the Student’s t-test. Values of P<0.05 were considered significant. Values of P<0.01 were considered very significant.
3. Results
3.1. Proteolytic activity of Lactobacillus bulgaricus
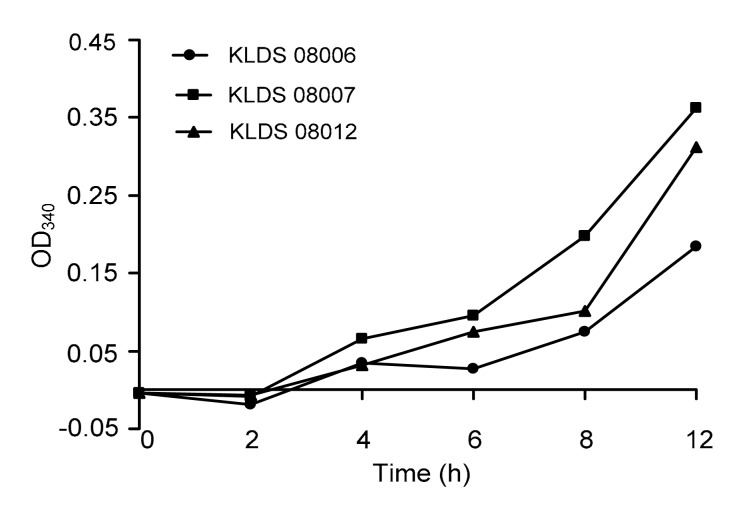
During fermentation, milk proteins were hydrolyzed by LAB proteinases and peptidases, resulting in an enhanced amount of free amino groups and peptides. Fig. 1 represents the proteolytic activities in RSM of three Lactobacillus bulgaricus strains (KLDS 08006, KLDS 08007, and KLDS 08012), which manifested the different activities of α-amino group release as a result of the proteolysis of milk proteins. The amount of liberated amino groups and peptides increased significantly during fermentation from 0 to 12 h for three strains (P<0.05). The extent of proteolysis in the KLDS 08007 fermented milk was significantly higher (P<0.01) than those of the KLDS 08012 and KLDS 08006 fermented strains in this study at the 12 h of culturing time. On the other hand, KLDS 08007 had the highest proteolytic activity in this study, which reached 0.363 of absorbance. The extent of proteolysis variance among strains appeared to be time-dependent.
Fig. 1.
Extent of proteolysis measured using OPA method during growth of three selected Lactobacillus bulgaricus strains in RSM for 12 h at 37 °C
3.2. Transcriptional expression of the target genes in Lactobacillus bulgaricus during different growth phases
In order to investigate how the proteinases, peptide transport proteins, and peptidases are regulated for peptide metabolism by Lactobacillus bulgaricus during different growth phases, we studied their expressions at the transcriptional level at the exponential growth and stationary phases. Total RNA was isolated at the exponential (6 h) and stationary (18 h) growth phases. Gene expression was determined by quantification of the respective transcripts in the cDNA library using 16S rRNA as the internal control by real-time PCR (Table 3). The gene expression decreased significantly with the growth of the strains. The transcription levels of the seven genes were reduced by 30-fold on average in the stationary phase compared with the exponential growth phase. Specifically, the transcription levels of the seven genes were reduced by 62.5-, 15.0-, and 59.0-fold in the strains KLDS 08006, KLDS 08007, and KLDS 08012, respectively, indicating that the expressions of the seven genes were significantly different among strains.
Table 3.
Expression of the genes related to peptide transport and hydrolysis in Lactobacillus bulgaricus during growth in MRS medium
| Growth phase | Strain number | Relative expression |
||||||
| OppD | PrtB | PepC | PepF | PepQ | PepX | PepT | ||
| Exponential | KLDS 08006 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| KLDS 08007 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| KLDS 08012 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Stationary | KLDS 08006 | 0.010±0.003** | 0.004±0.001** | 0.012±0.004** | 0.033±0.017** | 0.025±0.003** | 0.009±0.001** | 0.020±0.001** |
| KLDS 08007 | 0.036±0.002** | 0.319±0.069** | 0.008±0.002** | 0.010±0.004** | 0.004±0.001** | 0.006±0.001** | 0.087±0.003** | |
| KLDS 08012 | 0.012±0.006** | 0.028±0.015** | 0.004±0.001** | 0.006±0.003** | 0.011±0.004** | 0.004±0.002** | 0.053±0.042** | |
P<0.01, compared to the control group (exponential phase)
Data are expressed as mean±SD of triplicates
The gene expression was significantly increased with the growth of the strains, except the expression of the PepQ gene of the KLDS 08012 strain in milk (Table 3). Particularly, the transcription levels of the PepF and PepT genes were significantly up-regulated compared with the other five genes. Therefore, LAB depend on a proteolytic system to degrade milk proteins for their growth.
3.3. Transcriptional expression of the target genes in Lactobacillus bulgaricus cultivated at different initial pH values
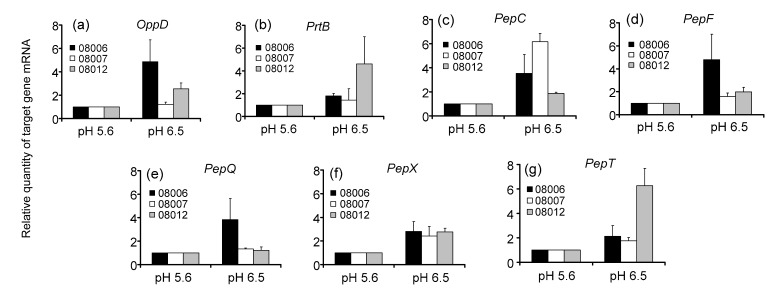
Total RNAs of Lactobacillus bulgaricus cultivated in normal MRS medium for 12 h at the initial pH of 5.6 and 6.5 were extracted to determine the gene expression by real-time PCR. The results are shown in Fig. 2. The expressions of the seven genes were up-regulated at initial pH 6.5 compared with that at initial pH 5.6. Furthermore, the change of gene expression was significantly different among different strains. In particular, expressions of OppD, PepC, PepF, PepQ, and PepX most significantly increased in strain KLDS 08006 at the initial pH 6.5; however, it is the expressions of PepC and PepX in strain KLDS 08007, and those of OppD, PrtB, PepX, and PepT in strain KLDS 08012 that were most significantly increased, compared with the initial pH 5.6.
Fig. 2.
Transcriptional expressions of the seven genes in Lactobacillus bulgaricus cells after cultivation at the initial pH of 5.6 and 6.5 for 12 h by real-time PCR analysis
(a) OppD; (b) PrtB; (c) PepC; (d) PepF; (e) PepQ; (f) PepX; (g) PepT. 16S rRNA was used as an internal control. Data are expressed as mean±SD of triplicates
3.4. Transcriptional expression of the target genes in Lactobacillus bulgaricus cultivated at different temperatures
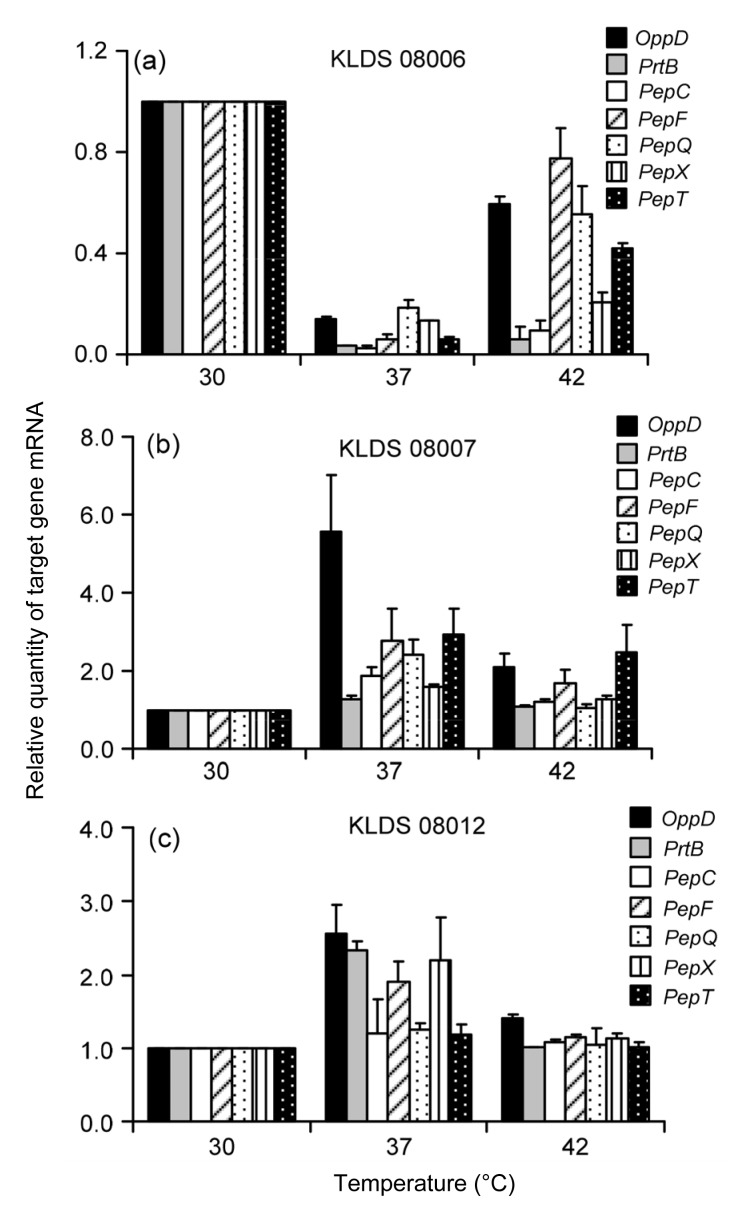
The transcriptional expressions of seven target genes in Lactobacillus bulgaricus cultivated in normal MRS medium for 12 h at 30, 37, and 42 °C, respectively, are shown in Fig. 3. The expressions of the key enzymes showed significant differences among the test strains cultured at the 37 °C for 12 h in normal MRS medium. The expressions of seven target genes of KLDS 08006 were the highest at 30 °C, which were significantly lower at 37 and 42 °C than at 30 °C (Fig. 3a). The expression of each target gene in KLDS 08007 and KLDS 08012 increased at first and then decreased, reaching the highest values at 37 °C (Figs. 3b–3c).
Fig. 3.
Transcriptional expressions of the seven genes in Lactobacillus bulgaricus after cultivation at 30, 37, and 42 °C for 12 h by real-time PCR analysis
(a) KLDS 08006; (b) KLDS 08007; (c) KLDS 08012. 16S rRNA was used as an internal control. Data are expressed as mean±SD of triplicates
3.5. Transcriptional expression of the target genes in Lactobacillus bulgaricus cultivated in MRS with different nitrogen sources
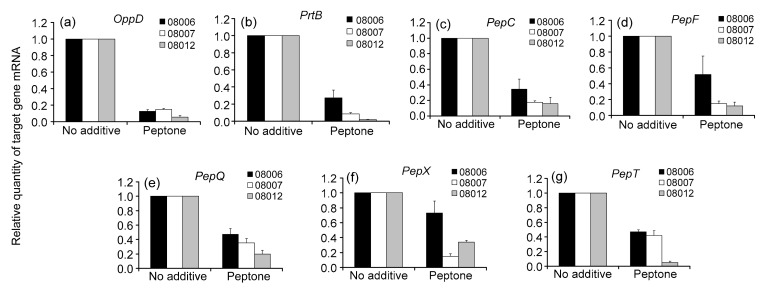
To evaluate the role of peptide source on the expression of genes involved in peptide uptake and hydrolysis, MRS medium containing extra peptone made from casein was used in addition to normal MRS. The total RNA was isolated after culturing in the normal MRS medium without additives and the MRS medium containing peptone for 12 h. As shown in Fig. 4, the transcription levels of the seven genes were reduced in the MRS medium containing peptone. The addition of the peptide source decreased the transcription levels of OppD, PrtB, PepC, PepF, PepQ, PepX, and PepT in exponentially growing cells on average by 9.1-, 8.0-, 4.4-, 3.8-, 2.9-, 2.5-, and 3.2-fold, respectively. Therefore, the effect of peptone supply on PepX transcription was the weakest compared with the other six genes (Fig. 4) and the impact on OppD transcription was the strongest. In addition, the expressions of the seven genes showed significant differences among the three strains (P<0.05). The expressions of all the seven genes decreased the most in strain KLDS 08012 and the least in strain KLDS 08006.
Fig. 4.
Transcriptional expressions of the seven genes in Lactobacillus bulgaricus cells after cultivation in the normal MRS without additives or the MRS containing peptone for 12 h by real-time PCR analysis
(a) OppD; (b) PrtB; (c) PepC; (d) PepF; (e) PepQ; (f) PepX; (g) PepT. 16S rRNA was used as an internal control. Data are expressed as mean±SD of triplicates
4. Discussion
4.1. Proteolytic activity of Lactobacillus bulgaricus
Only 1%–2% of milk proteins undergo proteolysis during milk fermentation and the principal milk protein is casein, but limited degradation of whey proteins may also occur (Szwajkowska et al., 2011; Griffiths and Tellez, 2013). Proteolysis leads to higher contents of peptides and free amino acids, especially histidine, valine, proline, and serine, in fermented milks than in non-fermented milk (Matar et al., 2003).
Production of amino acids and peptides from the degradation of milk proteins by LAB enzymes and utilization of these amino acids are central metabolic activities of LAB (Gobbettl et al., 2002). LAB, isolated from milk products, require 4 to 14 amino acids depending on the strain (Chopin, 1993). However, the amount of free amino acids and peptides in milk is very low. The amount of liberated amino groups and peptides increased significantly during fermentation from 0 to 12 h for three strains (P<0.05) in this study. These findings were consistent with those reported by Donkor et al. (2007).
4.2. Transcriptional expression of the target genes in Lactobacillus bulgaricus during different growth phases
Even though many studies have investigated the proteolytic ability of LAB, few reports determine the effect of growth media and different growth phases on their proteolytic system. It is a well-known fact that the proteolytic system of LAB is activated when milk is used as a growth medium (Wakai and Yamamoto, 2012). The results of this study indicated that the expression of key enzymes showed down-regulated trend in general in the MRS medium, but up-regulated trend in RSM in the stable phase compared with that in the logarithmic phase (the control group). These findings are consistent with those reported by Gitton et al. (2005) and Liu et al. (2012). Smeianov et al. (2007) reported that Lactobacillus helveticus CNRZ 32 genes encoding cell-envelope proteinases (CEPs), endopeptidases, and oligopeptide transporters were up-regulated when the strain was grown in milk compared with the levels of expression in the MRS medium. The CEP activities of six L. helveticus strains on αs1- and β-casein in MRS and milk were evaluated. It was shown that the CEPs of the different strains hydrolyzed intact αs1- and β-casein after growth in milk, but not in MRS (Jensen et al., 2009; Griffiths and Tellez, 2013). Hébert et al. (1997) showed that the effect of the growth medium on proteinase activity was evident with Lactobacillus helveticus CRL581. Smeianov et al. (2007) investigated the regulation of genes encoding peptidases of Lactobacillus helveticus CNRZ 32, and found that the PepI gene was up-regulated during growth in MRS medium, while the genes PepN, PepX, and PepR were up-regulated during growth in skim milk. The same level of expression of PepC was observed in skim milk and MRS. The expression of key enzyme genes decreased in the presence of an available nitrogen source and their activities increased in the growth medium of milk.
4.3. Transcriptional expression of the target genes in Lactobacillus bulgaricus cultivated under different conditions
Our results showed that the transcriptional expression of the genes in the proteolytic system was affected by the initial pH of the normal MRS medium, culturing temperature, the nitrogen source, and the growth phase. Moreover, the transcriptional expression of the genes was significantly different among different strains as the needs for amino acids were not the same among different strains (Neviani et al., 1995).
Real-time PCR analysis indicated that the gene expression was decreased significantly at the stationary phase and the extent of decrease was significantly different among different strains. Similar observations have been made by Vermeulen et al. (2005). The increased gene expression at the logarithmic phase is likely because more amino acids are needed for the fast growth at the logarithmic phase.
Sánchez et al. (2007) have observed higher expression of PepP in Bifidobacterium longum NCIMB 8809 during growth at pH 4.8 than at pH 7.0. However, there are few reports on the acid-induced changes in genes related to the proteolytic system in LAB. Our real-time PCR analysis confirmed the up-regulation of the genes for OppD, PrtB, PepC, PepF, PepQ, PepX, and PepT at the initial pH 6.5 compared with that at the initial pH 5.6. It has been observed that the strains grow faster in MRS at the initial pH 6.5 than at the initial pH 5.6. Therefore, the strains need more proteinases to produce more amino acids in order to maintain the fast growth. Wu et al. (2011) reported that the gene expression of the PepP was suppressed at the lower pH. The expression of all the studied genes demonstrated consistent changes with medium pH at the transcriptional level and the protein level (Argyle et al., 1976; Law, 1978).
Milk contains few amino acids and peptides and has low proteolytic activity. Therefore, proteinase activity is a prerequisite for the growth of LAB in milk. It is reported that Lactobacillus bulgaricus has stronger proteinase activity than Lactococcus lactis (Simova and Beshkova, 2007; Griffiths and Tellez, 2013). We found that the addition of peptides to MRS resulted in the reduced transcription of the seven genes studied in exponentially growing cells, which is consistent with previous reports on the biochemical and genetic studies in dairy Lactobacilli such as Lactobacillus helveticus and Lactobacillus delbrueckii subsp. lactis (Hébert et al., 2002) as well as Lactobacillus lactis (Guédon et al., 2001) that demonstrated sufficient peptides reduced the expression of proteinases. Vermeulen et al. (2005) reported that during the stationary phase, DtpT and Opp transcriptions are reduced regardless of the addition of peptides. Because amino acids and peptides are accumulated during fermentation, externally added peptides no longer affect the expression of peptide transporters during the stationary phase. Therefore, peptide availability strongly affects the expression of the genes in the proteolytic system.
5. Conclusions
In summary, the expressions of seven genes in the proteolytic system of Lactobacillus bulgaricus under different conditions were investigated. The gene expression significantly increased with the growth of the strains in RSM, except the PepQ gene of the KLDS 08012 strain. On the other hand, the gene expression was down-regulated in the MRS medium. We found that the initial pH of the MRS medium, nitrogen source, and growth phase affected the transcriptional expression of these genes. And the gene expressions of the different strains under different conditions were significantly different. Therefore, these factors must be taken into account for the effective use of this bacterium.
Footnotes
Project supported by the Program for New Century Excellent Talents in Heilongjiang Provincial University (No. 1253-NCEF-006), China
Compliance with ethics guidelines: Jun-cai HOU, Fei LIU, Da-xi REN, Wei-wei HAN, and Yue-ou DU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Argyle PJ, Mathson GE, Chandan RC. Production of cell-bound proteinase by Lactobacillus bulgaricus and its location in the bacterial cell. J Appl Microbiol. 1976;41(1):175–184. doi: 10.1111/j.1365-2672.1976.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 2.Azcarate-Peril MA, Tallon R, Klaenhamme TR. Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J Dairy Sci. 2009;92(3):870–886. doi: 10.3168/jds.2008-1457. [DOI] [PubMed] [Google Scholar]
- 3.Champomier-Vergès MC, Marceau A, Méra T, et al. The pepR gene of Lactobacillus sakei positively regulated by anaerobiosis at the transcriptional level. Appl Environ Microbiol. 2002;68(8):3873–3877. doi: 10.1128/AEM.68.8.3873-3877.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen MM, Li AL, Sun MC, et al. Optimization of the quenching method for metabolomics analysis of Lactobacillus bulgaricus . J Zhejiang Univ-Sci B (Biomed. & Biotechnol.) 2014;15(4):333–342. doi: 10.1631/jzus.B1300149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YS, Christensen JE, Broadbent JR, et al. Identification and characterization of Lactobacillus helveticus PepO2, an endopeptidase with post-proline specificity. Appl Environ Microbiol. 2003;69(2):1276–1282. doi: 10.1128/AEM.69.2.1276-1282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12(1-3):21–37. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 7.Christensen JE, Dudley EG, Pederson JA, et al. Peptidases and amino acid catabolism in lactic acid bacteria. Anton Leeuwenhoek. 1999;76(1-4):217–246. doi: 10.1023/A:1002001919720. [DOI] [PubMed] [Google Scholar]
- 8.Church FC, Swaisgood HE, Porter DH, et al. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci. 1983;66(6):1219–1227. doi: 10.3168/jds.S0022-0302(83)81926-2. [DOI] [Google Scholar]
- 9.Doeven MK, Kok J, Poolman B. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Mol Microbiol. 2005;57(3):640–649. doi: 10.1111/j.1365-2958.2005.04698.x. [DOI] [PubMed] [Google Scholar]
- 10.Donkor ON, Henriksson A, Vasiljevic T, et al. Proteolytic activity of dairy lactic acid bacteria and probiotics as determinant of growth and in vitro angiotensin-converting enzyme inhibitory activity in fermented milk. Lait. 2007;87(1):21–38. doi: 10.1051/lait:2006023. [DOI] [Google Scholar]
- 11.Genay M, Sadat L, Gagnaire V, et al. prtH2, not prtH, is the ubiquitous cell wall proteinase gene in Lactobacillus helveticus. Appl Environ Microbiol. 2009;75(10):3238–3249. doi: 10.1128/AEM.02395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert C, Atlan D, Blanc B, et al. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbruekii subsp. bulgaricus . J Bacteriol. 1996;178(11):3059–3065. doi: 10.1128/jb.178.11.3059-3065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitton C, Meyrand M, Wang J, et al. Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk. Appl Environ Microbiol. 2005;71(11):7152–7163. doi: 10.1128/AEM.71.11.7152-7163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobbettl M, Stepaniak L, de Angelis M, et al. Latent bioactive peptides in milk proteins: proteolytic activation and significance in dairy processing. Crit Rev Food Sci Nutr. 2002;42(3):223–239. doi: 10.1080/10408690290825538. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths MW, Tellez AM. Lactobacillus helveticus: the proteolytic system. Front Microbiol. 2013;4:30. doi: 10.3389/fmicb.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guédon E, Renault P, Ehrlich SD, et al. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J Bacteriol. 2001;183(12):3614–3622. doi: 10.1128/JB.183.12.3614-3622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hébert EM, Raya RR, de Giori GS. Characterization of a cell membrane-associated proteinase from Lactobacillus helveticus CRL 581. Curr Microbiol. 1997;35(3):161–164. doi: 10.1007/s002849900231. [DOI] [Google Scholar]
- 18.Hébert EM, Raya RR, de Giori GS. Modulation of the cell-surface proteinase activity of thermophilic lactobacilli by the peptide supply. Curr Microbiol. 2002;45(6):385–389. doi: 10.1007/s00284-002-3780-z. [DOI] [PubMed] [Google Scholar]
- 19.Jensen MP, Vogensen FK, Ardö Y. Variation in caseinolytic properties of six cheese related Lactobacillus helveticus strains. Int Dairy J. 2009;19(11):661–668. doi: 10.1016/j.idairyj.2009.04.001. [DOI] [Google Scholar]
- 20.Kunji ERS, Hagting A, de Vries CJ, et al. Transport of β-casein-derived peptides by the oligopeptide transport system is a crucial step in the proteolytic pathway of Lactococcus lactis . J Biol Chem. 1995;270(4):1569–1574. doi: 10.1074/jbc.270.4.1569. [DOI] [PubMed] [Google Scholar]
- 21.Kunji ERS, Mierau I, Hagfing A, et al. The proteolytic systems of lactic acid bacteria. Anton Leeuwenhoek. 1996;70(2-4):187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 22.Law BA. Peptide utilization by group N streptococci. J Gen Microbiol. 1978;105(1):113–118. doi: 10.1099/00221287-105-1-113. [DOI] [PubMed] [Google Scholar]
- 23.Liu E, Zheng H, Hao P, et al. A model of proteolysis and amino acid biosynthesis for Lactobacillus delbrueckii subsp. bulgaricus in whey. Curr Microbiol. 2012;65(6):742–751. doi: 10.1007/s00284-012-0214-4. [DOI] [PubMed] [Google Scholar]
- 24.Liu MJ, Bayjanov JR, Renckens B, et al. The proteolytic system of lactic acid bacteria revisited a genomic comparison. BMC Genomics. 2010;11:36. doi: 10.1186/1471-2164-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Matar C, LeBlanc JG, Martin L, et al. Biologically active peptides released from fermented milk: role and functions. In: Farnworth ER, editor. Hand Book of Fermented Functional Foods. 1st Ed. Boca Raton, FL: CRC Press; 2003. pp. 177–201. [Google Scholar]
- 27.Mayo B, Kok J, Venema K. Molecular cloning and sequence analysis of the X-propyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. cremoris . Appl Environ Microbiol. 1991;57(1):38–44. doi: 10.1128/aem.57.1.38-44.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neviani E, Giraffa G, Brizzi A, et al. Amino acid requirements and peptidase activities of Streptococcus salivarius subsp. thermophilus . J Appl Microbiol. 1995;79(3):302–307. doi: 10.1111/j.1365-2672.1995.tb03141.x. [DOI] [PubMed] [Google Scholar]
- 29.Niesters HG. Quantitation of viral load using real-time amplification. Methods. 2001;25(4):419–429. doi: 10.1006/meth.2001.1264. [DOI] [PubMed] [Google Scholar]
- 30.Peltoniemi K, Vesanto E, Palva A. Genetic characterization of an oligopeptide transport system from Lactobacillus delbrueckii subsp. bulgaricus . Arch Microbiol. 2002;177(6):457–467. doi: 10.1007/s00203-002-0411-9. [DOI] [PubMed] [Google Scholar]
- 31.Picon A, García-Casado MA, Nuñez M. Proteolytic activities, peptide utilization and oligopeptide transport systems of wild Lactococcus lactis strains. Int Dairy J. 2010;20(3):156–162. doi: 10.1016/j.idairyj.2009.10.002. [DOI] [Google Scholar]
- 32.Roland JS. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Anton Leeuwenhoek. 1999;76(1-4):139–155. doi: 10.1007/978-94-017-2027-4_6. [DOI] [PubMed] [Google Scholar]
- 33.Sánchez B, Champomier-Vergès MC, Collado Mdel C, et al. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl Environ Microbiol. 2007;73(20):6450–6459. doi: 10.1128/AEM.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savijoki K, Palva A. Purification and molecular characterization of a tripeptidase (PepT) from Lactobacillus helveticus . Appl Environ Microbiol. 2000;66(2):794–800. doi: 10.1128/AEM.66.2.794-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savijoki K, Ingmer H, Varmanen P. Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol. 2006;71(4):394–406. doi: 10.1007/s00253-006-0427-1. [DOI] [PubMed] [Google Scholar]
- 36.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 37.Simova E, Beshkova D. Effect of growth phase and growth medium on peptidase activities of starter lactic acid bacteria. Lait. 2007;87(6):555–573. doi: 10.1051/lait:2007036. [DOI] [Google Scholar]
- 38.Smeianov VV, Wechter P, Broadbent JR, et al. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl Environ Microbiol. 2007;73(8):2661–2672. doi: 10.1128/AEM.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefanitsi D, Garel JR. A zinc-dependent proteinase from the cell wall of Lactobacillus delbrueckii subsp. bulgaricus . Lett Appl Microbiol. 1997;24(3):180–184. doi: 10.1046/j.1472-765X.1997.00376.x. [DOI] [PubMed] [Google Scholar]
- 40.Stucky K, Klein JR, Schüller A, et al. Cloning and DNA sequence analysis of pepQ, a prolidase gene from Lactobacillus delbrueckii subsp. lactis DSM7290 and partial characterization of its product. Mol Gen Genet. 1995;247(4):494–500. doi: 10.1007/BF00293152. [DOI] [PubMed] [Google Scholar]
- 41.Szwajkowska M, Wolanciuk A, Barłowska J, et al. Bovine milk proteins as the source of bioactive peptides influencing the consumers’ immune system—a review. Anim Sci Pap Rep. 2011;29(4):269–280. [Google Scholar]
- 42.Tynkkynen S, Buist G, Kunji E, et al. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis . J Bacteriol. 1993;175(23):7523–7532. doi: 10.1128/jb.175.23.7523-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulve VM, Monnet C, Valence F, et al. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis . J Appl Microbiol. 2008;105(5):1327–1333. doi: 10.1111/j.1365-2672.2008.03869.x. [DOI] [PubMed] [Google Scholar]
- 44.Vermeulen N, Pavlovic M, Ehrmann MA, et al. Functional characterization of the proteolytic system of Lactobacillus sanfranciscensis DSM 20451T during growth in sourdough. Appl Environ Microbiol. 2005;71(10):6260–6266. doi: 10.1128/AEM.71.10.6260-6266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakai T, Yamamoto N. Antihypertensive peptides specific to Lactobacillus helveticus fermented milk. In: Sammour RH, editor. Biotechnology—Molecular Studies and Novel Applications for Improved Quality of Human Life. Biochemistry, Genetics and Molecular Biology. InTech; 2012. pp. 159–172. [DOI] [Google Scholar]
- 46.Wang JC, Zhang WY, Zhong Z, et al. Transcriptome analysis of probiotic Lactobacillus casei Zhang during fermentation in soymilk. J Ind Microbiol Biotechnol. 2012;39(1):191–206. doi: 10.1007/s10295-011-1015-7. [DOI] [PubMed] [Google Scholar]
- 47.Wu R, Zhang WY, Sun TS, et al. Proteomic analysis of responses of a new probiotic bacterium Lactobacillus casei Zhang to low acid stress. Int J Food Microbiol. 2011;147(3):181–187. doi: 10.1016/j.ijfoodmicro.2011.04.003. [DOI] [PubMed] [Google Scholar]