Figure 3.
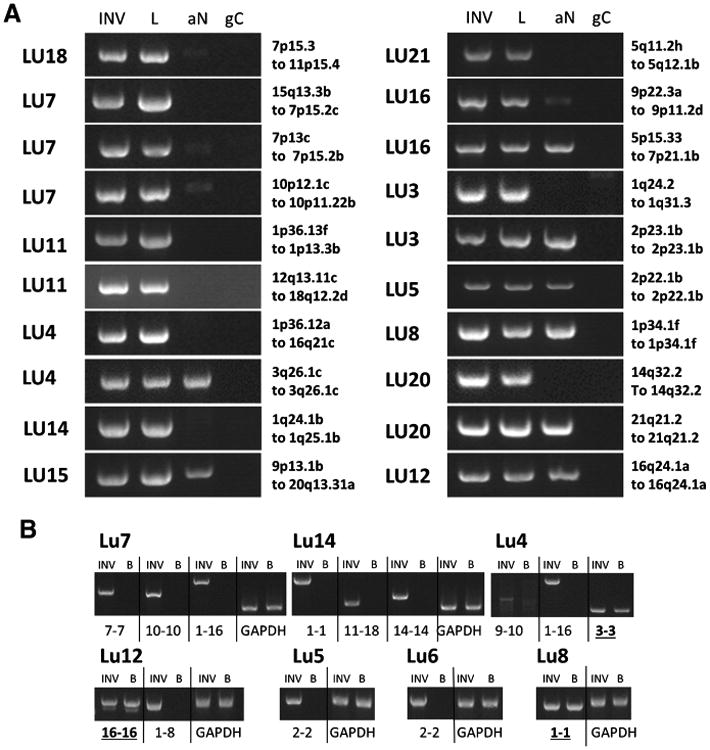
PCR validation of large genomic rearrangements. A, PCR products from primers flanking predicted genomic breakpoints for the invasive (INV) and lepidic (L) components, together with the associated normal (aN) and genomic control (gC). B, PCR validations on germline DNA for the indicated cases where blood was available and consented for that patient, compared with the associated lepidic tumor DNA. Case number and genetic loci of breakpoints or chromosomes involved are indicated; more precise genomic positions are presented in Supplementary Table S2. PCR products were run on 1.5% agarose gels together with 1 kb ladder (Ladder). Rearrangements present in blood are underlined and bold.
