Summary
Background
Circadian neural circuits generate near 24 hr physiological rhythms that can be entrained by light to coordinate animal physiology with daily solar cycles. To examine how a circadian circuit reorganizes its activity in response to light, we imaged period (per) clock gene cycling for up to 6 days at single neuron resolution in whole brain explant cultures prepared from per-luciferase transgenic flies. We compared cultures subjected to a phase-advancing light pulse (LP) to cultures maintained in darkness (DD).
Results
In DD, individual neuronal oscillators in all circadian subgroups are initially well synchronized, then show monotonic decrease in oscillator rhythm amplitude and synchrony with time. The s-LNvs and LNds exhibit this decrease at a slower relative rate. In contrast, the LP evokes a rapid loss of oscillator synchrony between and within most circadian neuronal subgroups followed by gradual phase retuning of whole circuit oscillator synchrony. The LNds maintain high rhythmic amplitude and synchrony following the LP along with the most rapid coherent phase advance. Immunocytochemical analysis of PER show these dynamics in DD and LP are recapitulated in vivo. Anatomically distinct circadian neuronal subgroups vary in their response to the LP, showing differences in the degree and kinetics of their loss, recovery and/or strengthening of synchrony and rhythmicity.
Conclusions
Transient desynchrony appears to be an integral feature of light response of the Drosophila multicellular circadian clock. Individual oscillators in different neuronal subgroups of the circadian circuit show distinct kinetic signatures of light response and phase retuning.
Introduction
Most organisms schedule their daily activity and metabolism using a circadian clock mechanism. Living organisms make daily adjustments to synchronize their circadian clock to seasonal changes of the 24-hr solar cycle by entrainment to environmental cues; light being the most powerful cue for most animals [1, 2]. The process of entrainment is most apparent when we travel rapidly across multiple time zones, i.e. jetlag. The brain circadian neural network of mammals is located in the suprachiasmatic nucleus (SCN), whereas the fruit fly Drosophila melanogaster and other insects have an anatomically distributed brain circadian neural circuit [3, 4]. Studies have revealed many similarities in the circadian biology of mammalian and Drosophila models, from molecular to circuit levels [5].
Longstanding efforts have been made to understand how clock cycling of individual neuronal oscillators distributed throughout circadian circuits maps to behaviors such as entrainment. Widely used immunocytochemical (ICC) analyses of rhythmic molecular clock components in circadian circuits are limited because they cannot capture individual oscillator longitudinal activity or dynamic relationships between oscillators in a single brain. The cross-sectional ICC approach takes individual “snap shots” of clock markers and requires averaging over many brains to construct an approximate time course. To circumvent these problems, longitudinal measurements of SCN oscillators have been made by multi-electrode recordings, or imaging of bioluminescent or fluorescent reporters of clock gene expression [6–8]. These studies reveal that individual SCN oscillators express a surprisingly large range of periods and phases. Further analysis of SCN oscillators has revealed how small molecule and peptide transmitters coordinate subsets of oscillators [5].
But the fundamental question of how a circadian network alters its distributed activity in response to a light entrainment signal in real time remains enigmatic. For the SCN, this is largely due to the technical difficulty of physiologically activating the melanopsin-mediated light input pathway in SCN slice cultures. Measuring the circuit-wide response to light is feasible in Drosophila because the entire fly brain can be cultured [9] and approximately half the neurons in the fly circadian circuit autonomously express the blue light receptor Cryptochrome (CRY) [10, 11], which provides the primary mechanism for light resetting the circadian clock and acute light evoked increases in firing rate in circadian neurons [12, 13]. To address how light reorganizes the activity of the Drosophila circadian circuit mapped at single cell resolution, we developed a culture system for Drosophila adult whole brains [9], then refined and combined high resolution imaging of circuit-wide single oscillators [14, 15] with sophisticated mathematical analytical tools [16, 17]. For in vivo comparison, we performed anti-PER ICC using the same light/dark protocols used for whole brain imaging. Although ICC has limited temporal resolution for single oscillator kinetics relative to bioluminescence recordings, we can test predictions of neuronal subgroup patterns of dynamic PER activity in response to light.
Results
Imaging the Drosophila circadian neural circuit in organotypically cultured whole adult brains prepared from XLG-Per-Luc flies
The Drosophila circadian circuit consists of at least six neuronal subgroups [18] which can be further subdivided by neurochemical or promoter fragment expression markers [19–23]. These include the large and small ventral lateral neurons (l-LNv and s-LNv), the dorsal lateral neurons (LNd), and three subgroups of dorsal neurons (DN 1, 2 and 3) (Figure S1A, DN2s not shown). The Drosophila circadian pacemaker neurons are functionally defined as cells that rhythmically express the clock proteins Period (PER) and Timeless (TIM).
Transgenic XLG-luc (XLG-Per-Luc) flies were used in this study because the 13.2 kb per gene promoter fragment drives expression of a PER-luciferase fusion protein in nearly all neurons of the circadian circuit. Normal behavioral rhythmicity is nearly restored when XLG-Per-Luc flies are crossed with the non-rhythmic per null mutant line per01 [24]. The spatiotemporal pattern of expression and degradation of the XLG-PER-LUC fusion protein resemble the native PER protein (Movie S1, Figure 1–6) [24]. Using a high quantum efficiency CCD camera, the anatomically defined major circadian neuron subgroups can be visualized by bioluminescence imaging of whole adult brains of XLG-Per-Luc flies (Figure S1C). Brains were maintained using a long-term organotypic culture protocol we developed in collaboration with the Hassan lab [9].
Figure 1. Oscillators in constant darkness demonstrate gradual desynchrony over time, whereas oscillators exposed to a white light pulse at CT 22 show synchrony phase retuning.
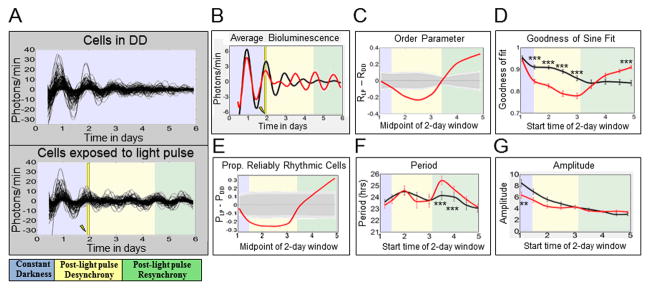
Neuronal oscillators were either maintained in constant darkness (‘DD cells’) or exposed to a 15 min 12.57 W/m2 (2,000 lux) light pulse at CT 22 on the second day in DD (‘LP cells’). The time at which the light pulse (LP) is applied is indicated by a yellow bar and lightning bolt. The colored-backgrounds provide general time frames of significant changes in order parameter. Bluish-gray indicates pre-LP application, yellow indicates post-LP desynchrony, and green indicates resynchrony. A: XLG-Per-Luc bioluminescence time-series measurements show that LP cells (lower panel; n=126) exhibit transient loss then recovery and even strengthening of cell synchrony over time compared to DD cells (upper panel; n=122), which exhibit a gradual, monotonic loss of cell synchrony. B: Comparing averaged bioluminescence traces confirms that LP cells (red line) exhibit an acute decrease in synchronized rhythmicity after the light pulse followed by recovery and eventual strengthening of synchronized rhythmicity relative to DD cells (black line). C: After a LP, oscillators display significant reduction in the order parameter R followed by a delayed significant increase in R. The order parameter R varies between 0 and 1, with higher values indicating similarity in phase, period, and waveform. The solid red curve represents the difference in R between LP and DD cells (RLP – RDD). The dark and light gray zones indicate the 95% and 99% confidence zones, respectively. The null hypothesis is that there is no difference between LP and DD values of R, as determined using 10,000 bootstrap samples (details in “Experimental Procedures”). D: Using oscillator goodness-of-sine-fit (g.o.f.) as a measure of rhythmicity, it was found that after a LP, cells (red line) demonstrate an acute reduction in g.o.f. followed by significantly greater g.o.f. over time as compared to DD oscillators at corresponding time points (black line). E: After a LP, relative to DD, there is a significant transient decrease in the proportion of reliably rhythmic cells (‘P’), followed by a significant increase in ‘P’ over time. The solid red line indicates the difference between LP and DD conditions (PLP – PDD). Cells with g.o.f. ≥ 0.82 are considered to be “reliably rhythmic.” The dark and light gray zones indicate 95% and 99% confidence zones as described in C. F: Sine-fit estimates of period indicate that LP cells (red line) exhibit a transient increase in period length several days after a light pulse. It should be noted that sine-fit estimates of period at these time points may be unreliable due to low amplitude oscillations following the light pulse. G: Sine-fit estimates of amplitude indicate that that LP cells (red lines) exhibit no significant differences in amplitude following exposure to the LP when compared to DD cells at corresponding time points. The difference in amplitude for the first 2-day window time point is likely due to slight overlap with changes in amplitude induced by the light pulse at 1.92 days. The error bars for g.o.f., period and amplitude represent ± SEM with significance analyzed using one-way ANOVA, Tukey post hoc test. *** indicates P<0.001 and ** indicates P<0.005.
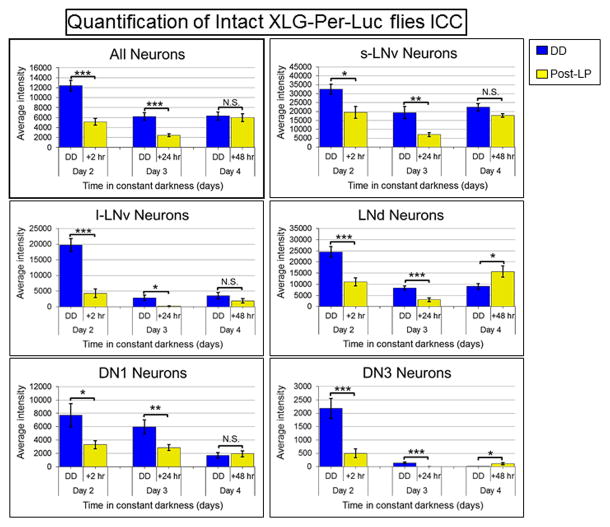
Figure 6. Quantification of significant changes in PER staining intensity from whole brains of XLG-Per-Luc flies either maintained in DD or exposed to a light pulse in vivo.
The software Volocity (PerkinElmer) was used to measure the average fluorescence intensity of PER staining in individual neurons visualized qualitatively in Figure 5. Neuronal oscillators in DD (blue) generally exhibit a gradual reduction in the average intensity of PER staining over time with the s-LNv showing the most stable amplitude. Conversely, neuronal oscillators exposed to a light pulse (LP, yellow) exhibit a significant reduction in PER staining intensity 24 hours after the LP and a significant recovery of staining intensity 48 hours after the LP. The LNds and DN3s even appear to exhibit a significant increase in PER staining intensity 48 hours after the LP in comparison to corresponding neurons maintained in DD. However, it should be noted that very dim fluorescence at later time points and tight clustering makes analysis of DN3s difficult. The error bars represent ± SEM. N.S. indicates no significant difference, * indicates P<0.05, ** indicates P<0.005 and *** indicates P<0.001 determined using Student’s t-test. The Student’s t-test was used to compare corresponding DD and LP neuronal oscillators with the null hypothesis that there is no difference in average PER staining fluorescence intensity. The laser intensity and other settings were kept the same for all groups for comparison of fluorescence intensities.
A phase-advancing light pulse induces acute desynchrony of most oscillators throughout the Drosophila circadian circuit followed by gradual phase retuning of synchrony
To determine the baseline circuit-wide dynamic relationship between individual oscillators, we imaged whole adult brains of XLG-Per-Luc flies (previously entrained under 12:12h LD, [24]) to measure single neuron oscillations in constant darkness (DD) for six days in organotypic culture [9]. Time series analyses of single neuron bioluminescence oscillations for ‘all DD cells’ (from all circadian neuronal subgroups, n = 122) in continuous 6 day DD recordings show initially synchronized oscillators throughout the circadian circuit that gradually decrease their oscillator amplitude and desynchronize with time, as shown by superimposed single-cell oscillator traces (Figure 1A, upper panel), averaged record (Figure 1B, black trace), and goodness-of-sine-fit as a measure of rhythmicity (Figure 1D, black trace). Average oscillator period is initially close to 24h for the first several days in DD, then decreases (Figure 1F, black trace). Oscillator amplitude decreases for all cells in DD, the s-LNvs dampen at a slower rate (Figure 1G (black trace), in agreement with whole animal and whole brain bioluminescence measurements of XLG-Per-Luc flies [24].
Next, we imaged the circadian network response in adult cultured whole brains prepared from XLG-Per-Luc flies exposed ex vivo to a phase-advancing white light pulse (LP) at CT 22 of the second day of DD (6 days total recording). We compared the circadian circuit dynamics for the LP response of individual oscillators relative to control baseline measurements for corresponding oscillators in DD at matched time points. In contrast to DD conditions, the LP evokes rapid desynchrony of oscillator cycling followed by gradual recovery then strengthening of synchrony 1–2 days after the LP that can be seen qualitatively in superimposed individual oscillator traces (Figure 1A, lower panel) and in the averaged record (Figure 1B, red trace). We call the entire dynamic process of gradual emergence of phase-shifted, high amplitude, and tighter synchrony oscillations following transient phase desynchrony after exposure to the phase advancing LP ”phase retuning”. The qualitatively similar phenomenon of transient phase desynchrony in SCN slices in response to bath applied vasoactive intestinal peptide (VIP) has been referred to as “phase tumbling” [25]. Examination of the detrended traces and the averaged traces for LP cells in Figure S2 (bottom) clearly demonstrates that cells exposed to the LP exhibit greater synchrony and phase-shifted rhythmicity at the end of the recording relative to cells in DD. To quantify order parameter R as a measure of the dynamic response of oscillator synchrony, we calculated values of R for a sequence of 2-day sliding windows using the definition of order parameter in [26]. R can range from 0 to 1 with higher values indicating similarity in phase, period and waveform. RLP – RDD was then calculated for all matched time points in the LP and DD datasets. Following the light pulse, we measure significantly negative values (RLP – RDD < 0) as ‘desynchrony’, subsequent values with no significant difference between the conditions (RLP – RDD ≈ 0) as ‘recovery,’ and significantly positive values (RLP – RDD > 0) at the end of the recordings as ‘strengthened.’ Overall analysis of ‘all LP cells’ (i.e. from all neuronal subgroups, n = 126) shows rapid and significant oscillator desynchrony relative to DD immediately following the LP (Figure 1C, yellow shaded area) which slowly phase retunes, with significantly strengthened oscillator synchrony by 2–3 days following the LP (Figure 1C, green shaded area). Analysis of goodness-of-sine-fit (g.o.f.) as a measure of rhythmicity over 2-day sliding windows yields a similar pattern of results: acute LP-reduced g.o.f. (Figure 1D, yellow shaded area) followed by gradual strengthening of oscillator g.o.f. several days later (Figure 1D, green shaded area). To confirm these patterns, we measured dynamic changes in the proportion of reliably rhythmic cells (PLP – PDD). The same trends of significant decreases in response to the LP relative to DD followed by recovery over several days are shown (Fig 1E). The periods of DD and LP cells are comparable and relatively stable with the exception of two later time points (Figure 1F). The overall amplitude of single-cell oscillators declines monotonically and does not differ significantly between LP and DD oscillators at time points following the light pulse (Figure 1G). Thus, changes in oscillator synchrony and phase form the major qualitative and quantitative response to light.
Neuronal subgroups exhibit qualitatively apparent differences in dynamics of PER activity both in DD and in response to a phase-advancing light pulse
We then longitudinally measured PER expression rhythms in single neurons from defined circadian neuronal subgroups in bioluminescence images collected at 30 min intervals for six days in DD from cultured whole adult brains of XLG-Per-Luc flies. The s-LNvs show the most robust rhythms and greatest inter-neuronal synchrony in DD compared with other subgroups (Figure 2A, top). The l-LNv also exhibit relatively large amplitude and coherent rhythms in DD, though to a lesser extent than the s-LNvs (Figure 2A, top). Previous reports on l-LNv oscillations dampening in DD yielded different conclusions. Some report l-LNv oscillations dampening within the first two days in DD [27, 28], while other studies report measurable l-LNv cycling of per mRNA after 9 days in DD [29] and protein levels [30] for at least 2.5 days in DD. We have reported considerably longer PER cycling (albeit out of phase) and phasic electrical circadian rhythmicity in the l-LNv after 15 days of DD by calibrating data collection time points to behavioral landmarks for each fly tested [31, 33]. Thus our present bioluminescence results support the findings in [29–31, 33]. The LNds, DN1s and DN3s show somewhat less robust rhythms with patterns of dampening amplitude and gradual loss of coherent rhythms over the six days of DD (Figure 2A, top).
Figure 2. Exposure of cultured brain explants to a light pulse reveals qualitatively distinct dynamic signatures of neuronal subgroups.
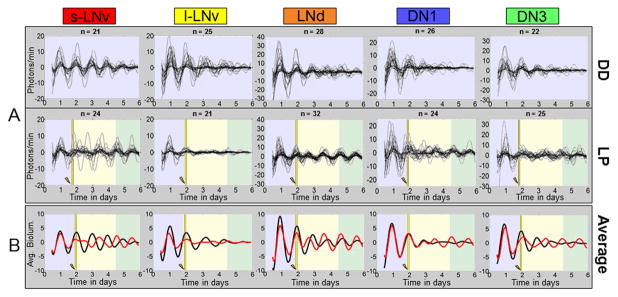
A: Single neuron oscillations are shown separately for each neuronal subgroup. Top: Neuron subgroups maintained in DD showing a general loss of intra-subgroup synchrony and amplitude over time. S-LNvs exhibit the most robust rhythms over time. Bottom: Neuron subgroups exposed to a 15 minute 12.57 W/m2 light pulse at CT 22 of the second day in DD ex vivo. LP induced transient phase tumbling followed by synchrony phase retuning is seen qualitatively at varying degrees for all groups except the l-LNv which rapidly lose oscillator synchrony and amplitude and do not phase retune following the LP by the end of the recording. Conversely, LNds do not appear to exhibit any significant loss of synchrony following the LP. The number of cells analyzed for each group is indicated by “n.” The background color coding is the same as in Fig. 2. B: Averaged bioluminescence traces for LP (red line) vs. DD (black line) oscillators sharpen the qualitative patterns seen in the individual oscillator records.
Single neuron oscillators from the defined circadian neuronal subgroups exposed to a LP show strikingly different dynamics compared to DD (Figure 2A, bottom). The s-LNv oscillations initially show coherent, high rhythm amplitudes similar to the DD condition, then exhibit marked desynchrony immediately after the LP, followed by a gradual recovery that phase retunes to shifted synchrony after several days (Fig 2A, bottom). In contrast, the l-LNvs exhibit immediate dampening of amplitude and weak rhythmicity following the LP that does not recover (Figure 2A, bottom). Of all the circadian neuronal subgroups measured, the l-LNvs appear to have the most labile and immediate response to the LP, consistent with previous findings as being light sensitive [13, 32–34]. In contrast, the LNds appear to maintain surprisingly high amplitude rhythms and coherence even after the LP. The DN1 and DN3 oscillators both show desynchronization, followed by recovery of synchrony several days after the LP (Figure 2A). The averaged traces for each circadian neuronal subgroup (Figure 2B) sharpen the qualitative assessments of single-cell traces for each condition. Averaged LNd oscillations show a remarkable immediate shift to an earlier phase in response to the phase-advancing LP without loss of amplitude relative to the DD condition.
Different circadian neuronal subgroups exhibit quantitatively distinct kinetic signatures for both DD and LP oscillator patterns
We analyzed each of the subgroups for their single-cell order parameters, g.o.f., and proportion of reliably rhythmic cells; comparing LP relative to DD. As a measure of synchrony over time among cells within a subgroup, the order parameter R was calculated for a sequence of 2-day sliding windows (Figure 3A). The s-LNvs show a significant loss of oscillator synchrony in response to the LP followed by gradual recovery (RLP – RDD ≈ 0) several days after the LP. The DN3 also show a significant loss of synchrony in response to the LP, but with a slower onset and more rapid recovery relative to the s-LNvs. In contrast to the s-LNv, no significant differences in R are seen for light-evoked l-LNvs relative to the DD baseline. The LNds and DN1s show significant increases in R coinciding with s-LNv recovery several days after the LP with the LNds exhibiting the earliest and greatest strengthening of synchrony relative to DD baseline values.
Figure 3. Neuronal subgroups respond to a phase-advancing light pulse with quantitatively distinct dynamics of transient desynchrony followed by recovery and strengthening of synchrony and rhythmicity.
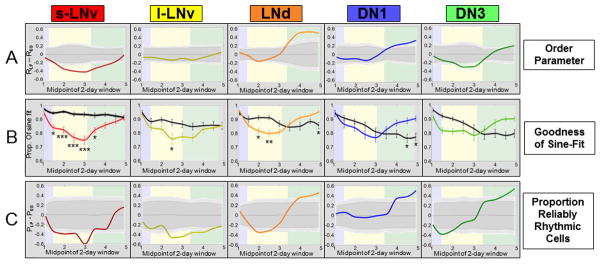
Colored-background frames of reference are the same as seen in Fig. 1. Circadian parameters are measured over 2 day sliding windows. A: After a light pulse (LP), neuronal subgroups exhibit transient loss and/or subsequent gain of synchrony with varying degrees and kinetics of response (s-LNv, LNd, DN1, DN3) or no significant response (l-LNv). Solid lines represent the difference in R between LP and DD conditions (RLP – RDD). Dark and light gray zones indicate 95% and 99% confidence intervals, assuming the null hypothesis of no difference between LP and DD. B: Exposure to LP results in a significant rapid reduction in the goodness-of-sine-fit (g.o.f.) for the s-LNvs, LNds and l-LNvs (listed by order of response). The DN1s and LNds demonstrate strengthened g.o.f. delayed by several days after the light pulse. Colored lines indicate average values for g.o.f. for LP cells, whereas solid black lines indicate values for DD cells. Error bars represent ± SEM. Significant differences between LP and DD conditions at each time point are indicated by *** for P<0.001, ** for P<0.005, and * for P<0.05 (one-way ANOVA, Tukey post hoc test). C: Analysis of the proportion of reliably rhythmic cells after a LP relative to the DD condition (PLP – PDD) reveals a significant initial decrease for the s-LNvs and l-LNvs and, to a lesser extent, the LNds and DN3s. The LNds, DN1s and DN3s demonstrate a later increase in proportion of reliably rhythmic cells compared to corresponding neurons in DD. Confidence intervals are plotted as described above.
Analysis of g.o.f. as an independent measure of rhythmicity for each neuronal subgroup supports the conclusions as determined by changes in the order parameter R in response to the LP (Figure 3B). The s-LNvs, LNds, and l-LNvs show significant decreases in g.o.f in response to the LP ranked as listed. The LNds and DN1s exhibit a significant but delayed increase in g.o.f several days after the LP. The DN3 exhibit a general trend of transient reduction followed by an increase in g.o.f though without reaching a significant difference between LP and DD. For proportion of reliably rhythmic cells (Figure 3C), the s-LNvs show significant decreases initially following the LP, as do the l-LNvs to a lesser extent, while the LNd, DN1 and DN3 subgroups show delayed significant increases that correspond to their phase retuning of synchrony. Thus, loss and subsequent recovery and/or strengthening of synchrony are quantifiable features of the circadian network’s response to phase advancing light that vary in a stereotypic manner between circadian neuron subgroups.
We also employed BPENS (Bayesian parameter estimation for noisy sinusoids) calculations over 2-day sliding windows as previously described [16] to quantify confidence in our criterion for reliably rhythmic cells and sine-fit estimates of periods (Figure S3). BPENS calculations confirmed the same distinct trends of light response for ‘all cells’ and for each neuronal subgroup (see Supplemental Information for details). Additionally, we ran a test using surrogate data from [16] using 2-day windows to further validate the accuracy of the sine-fit measures with wavelet-detrending method that we employed. The resulting period estimates had a mean absolute error of 1.6% with a standard deviation of 1.2%. This test, along with the BPENS correlation measures, confirms that the quantified trends in light response are consistent and reliable.
Circadian neuronal subgroups respond to the LP with temporally distinct kinetic signatures of transient desynchrony followed by phase retuned synchrony
Under DD conditions, the different circadian neuronal subgroups are initially synchronous, but gradually decrease their inter-group synchrony over six days as seen in the aligned averaged per gene driven bioluminescence signals (Figure 4A, top panel). Surprisingly, given the proposed role of the s-LNv as ‘master oscillators’, the averaged peaks of the DN1s, DN3s and LNds temporally lead the lateral s-LNv and l-LNv in DD (Figure 4A, top panel). This may be due to shorter free-running periods in these neurons as proposed in [35] that circadian periods may be established by synergistic interactions between multiple neuronal subgroups rather than encoded by a single neuronal subgroup like the s-LNvs. The LP induces acute desynchrony between the circadian subgroups, shown by the aligned averaged per driven bioluminescence signal peaks, followed by phase retuning of synchrony that varies between circadian subgroups after the LP (Figure 4A, lower panel). Comparison of the order parameter R within each cell subgroup shows the same temporal sequence described above of significant light-induced acute desynchrony followed several days later by significant strengthening of oscillator synchrony (Figure 4B). This distributed dynamic pattern of light response is similar for the proportion of reliably rhythmic cells (Figure 4C). Comparative dynamic spatiotemporal patterns are depicted in Movie S2 with individual frames in Figure 4D, in which the values of R for each neuronal subgroup are converted to a color heat map (DD on the left and LP on the right).
Figure 4. Alignment of neuronal subgroup responses to a LP reveals temporally distinct kinetic signatures of phase retuning.
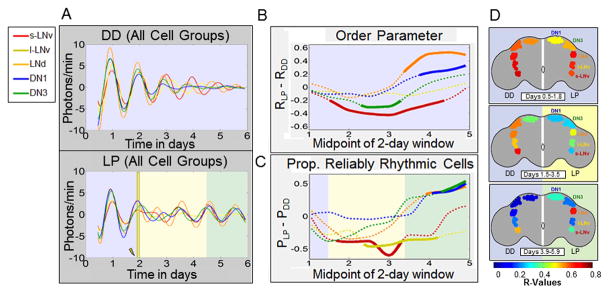
In A–C, plots of neuronal subgroup data are coded by color: s-LNv (red), l-LNv (yellow), LNd (orange), DN1 (blue) and DN3 (green). A (Top): Average bioluminescence traces for subgroups maintained in DD exhibit a progressive and monotonic loss of rhythmicity and inter-subgroup synchrony over time. A (Bottom): After a light pulse (LP), average bioluminescence traces for subgroups exhibit a transient reduction in rhythmic amplitude and inter-subgroup synchrony, followed by a general strengthening of rhythmic amplitude and inter-subgroup synchrony over time relative to corresponding neurons in DD. B, C: Inter-subgroup comparisons of averaged single neuron circadian parameters measured using 2 day sliding windows. B: After a LP, s-LNvs exhibit the first and longest lasting significant reduction in R, with DN3s exhibiting similar but less extreme changes. LNds and DN1s subsequently show significant strengthening of synchrony, coinciding with recovery of s-LNv synchrony. Dotted lines indicate no significant changes in synchrony after a light pulse relative to DD (RLP – RDD) while solid lines indicate significance outside the 99% confidence interval determined by bootstrapping. C: Inter-subgroup comparisons of the relative proportion of reliably rhythmic cells (PLP – PDD) show that the s-LNvs and l-LNvs exhibit significant initial decreases in proportion of rhythmic cells after exposure to a light pulse, whereas the DN1, DN3 and LNd exhibit a significant delayed increase. Dotted and solid lines indicate lack or presence of statistically significant differences between LP and DD conditions as shown above for R. D: Images of selected time points from Movie S2 comparing inter-subgroup differences in kinetics of changes in synchrony in DD or with the light pulse. The pseudocolor heat map codes values of R, with warm colors indicating high synchrony among cells within a subgroup. Left sides of brains show DD, right sides show response to LP. The colored backgrounds designating general time frames of significant changes in R are the same as previous figures.
Adult XLG-Per-Luc flies exposed to a light pulse in vivo exhibit transient reduction followed by delayed increase in PER staining intensity relative to DD
After observing dynamic changes in PER activity in whole brain explants exposed to a light pulse (LP), we predicted that the same trends of light-induced network desynchrony and resynchrony would be observed for neuronal subgroups in the brains of adult, male XLG-Per-Luc flies exposed to a light pulse in vivo. Accordingly, we adapted the DD and LP protocols in vivo followed by brain collection for anti-PER ICC analysis of individual neuronal oscillator PER activity. Whole brains in DD were fixed near expected daily peaks of PER based on previous entrainment history. Whole brains of flies exposed to the LP were fixed at projected daily peaks of PER based on the expected phase advance by the LP (see experimental procedures for details).
In Figure 5, neuronal subgroups are stained for PER (green) and PDF (red) from standardized laser and imaging settings (‘std gain’) for relative comparison of staining intensity along with higher gain (‘high gain’) settings optimized to compensate for later time points and dimmer neuronal subgroups (e.g. the DN3s). In line with their proposed role as key regulators of behavior in DD [27, 28, 36], the s-LNvs exhibit the greatest and most sustained PER staining intensities over time in DD. Uniformly contrasting DD baseline measures, oscillators exposed to a LP (labeled LP + number of hours since exposure, yellow background) show a decrease in PER staining intensity immediately after the light pulse (LP + 2 hours) with the most qualitatively apparent decrease 24 hours after the light pulse (Figure 5). 48 hours after application of the LP, most neuronal subgroups exhibit recovery of staining intensity – recovery for dimmer subgroups such as the DN1, DN3 and l-LNv is more distinct by quantitative measurements (see below). Remarkably, phase retuning is measurable by anti-PER ICC as the LNd exhibit a qualitatively distinct and statistically significant increase in PER staining 48 hours after LP exposure relative to LNds maintained in DD. Anti-PER ICC also show significantly higher levels of PER in the DN3 for LP day 4 at 48 hr post LP relative to day 3 at 24 hr post LP (Figures 5 and 6). The 4 day range of the in vivo ICC staining protocol shows that all of the major features of network transient desynchrony and synchrony phase retuning following a phase advance light pulse are shared between whole brain longitudinal per-luc imaging and in vivo.
Figure 5. Exposure of intact XLG-Per-Luc adult flies to a light pulse in vivo reveals qualitatively apparent transient loss and subsequent increase in PER staining intensity over time.
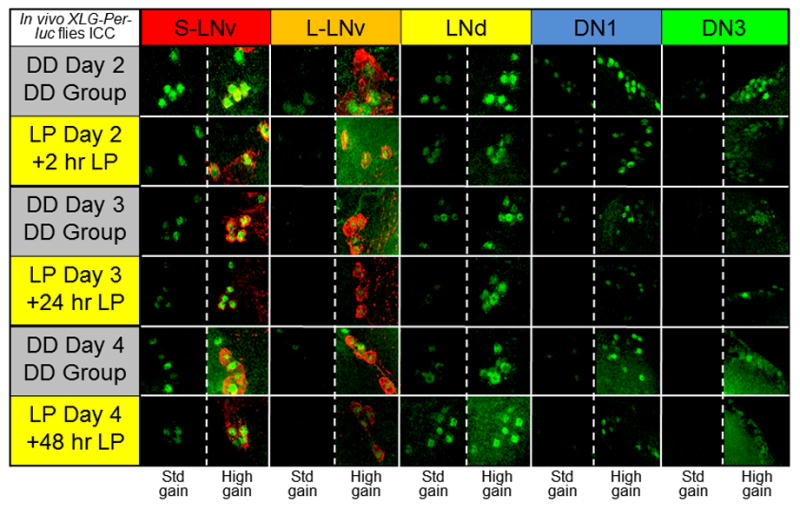
After entrainment to a standard 12h/12h LD schedule for 3 days, adult XLG-Per-Luc flies were either maintained in DD (‘DD group’; gray background) or exposed to 15 min 12.57 W/m2 (2,000 lux) light pulse at CT 22 on the second day in DD in vivo (labeled ‘LP + number of hours since exposure’; yellow background). Adult whole brains were stained for PER (green) and PDF (red). Flies in the DD group were fixed at CT 22 for DD day 2 and CT 0 for DD days 3 and 4. Flies exposed to the LP were fixed 2 hours (CT 2), 24 hours (CT 0), and 48 hours (CT 0) after the light pulse. Note that fixation times for LP flies are recalibrated such that the new CT 0 corresponds to the time when the LP is administered. In comparison to corresponding DD cells, it can be seen from representative ICC images that all neuronal subgroups demonstrate substantial dampening of PER staining intensity 24 hours after light pulse exposure with general recovery of amplitude 48 hours after the LP. The staining for each neuronal subgroup is presented at the same standardized (‘std gain’) laser and microscope settings to compare between time points and conditions along with staining obtained with higher intensity settings (‘high gain’) for visualization of dim fluorescence – particularly for later time points and the DN3s. See “Experimental Procedures” for details regarding ICC protocol and fixation times.
Neuronal subgroups exposed to a light pulse in vivo exhibit quantitatively distinct and significant changes in PER staining relative to corresponding oscillators in DD
In Figure 6, quantification of average PER fluorescence intensity for oscillators exposed to a phase advancing white light pulse in vivo reveals similar trends in phase retuning observed in our bioluminescence recordings with brain explants exposed to a light pulse ex vivo. Relative to baseline measurements of PER staining intensity for ‘all neurons’ in DD (averaged from all neuronal subgroups, blue), ‘all neurons’ exposed to the LP (yellow) exhibited a global significant reduction in staining intensity within 2 hours of light exposure with the decrease in intensity continuing even up to 24 hours after the LP. 48 hours after the light pulse, the PER staining intensity has generally recovered (i.e. no significant difference in intensity between LP and DD oscillators). This general recovery of staining intensity 2 hours in advance of the original peak indicates a network phase shift induced by the phase advancing light pulse. The s-LNvs, l-LNvs and DN1s exhibit this trend to varying degrees. Furthermore, the LNds and DN3s exhibit a significant increase in PER staining intensity 48 hours after exposure to the LP relative to corresponding oscillators in DD.
In vivo ICC experiments repeated for adult w1118 flies show the same trends of PER activity in DD and in response to phase advancing LP as XLG-Per-Luc ICC (Figure S4-S6). Quantitative comparison of PER levels between w1118 (red) and XLG-Per-Luc (violet) flies show no significant difference in staining intensity between corresponding neurons between matched conditions and time points (Figure S4). The similarity of PER staining intensities between w1118 and XLG-Per-Luc flies supports previous studies [24, 37] indicating that XLG-Per-Luc flies are a reliable model to study dynamics of PER activity. The common trend of transient loss then recovery and/or strengthening of PER staining intensities at expected phase-shifted peak times relative to expected peak intensities in DD provides further evidence that LP-induced transient desynchrony and delayed synchrony phase retuning observed in cultured brain explants is recapitulated in vivo (Figure 5–6).
Discussion
Multi-day functional imaging of organotypic cultures of Drosophila whole adult brains requires long term health of the cultures. Our previous work shows that cultures maintain identifiable morphological characteristics of the LNvs for up to 20 days and TIM clock protein cycling identified by ICC in single LNv up to 3 days [9]. We now reliably measure longitudinal circuit-wide function of single neuron oscillators by XLG-Per-Luc bioluminescence up to six days. The minimal Drosophila circadian network of six neuronal subgroups can be further subdivided based on neurochemical or genetic markers [21–26]. The current study is restricted to characterizing the general dynamic activity of the classical anatomically recognized s-LNv, l-LNv, LNd, DN1 and DN3 subgroups, which show distinct kinetic signatures in DD and in response to a phase advancing LP. Future studies will parse other divisions of the circuit.
The whole brain cultures tend to flatten with time, causing slight gradual positional distortion of the circadian neurons which actually makes for easier identification and isolation of single neuronal oscillators – particularly for dense subgroups such as the DN3s. We employed rigorous criteria. Oscillators that could not be clearly anatomically identified, isolated from nearby cells, distinguished from frame to frame, and did not exhibit cycling throughout the recordings were excluded from analysis. DN3 neurons do not express the CRY photoreceptor, require signaling from CRY-positive neurons to respond to light. Thus their LP response shows the circadian neural circuit remains intact in cultures [10, 11]. Intact flies can also light entrain via rhodopsin-based photic input from the eyes and other external photoreceptors [12]. We exclude photoreceptors from cultures as they increase the risk of microbiological contamination. Glass60j mutant flies that lack all external photoreceptors retain light responsiveness, normal behavioral entrainment and PER cycling (ICC) in a CRY-dependent manner [12]. We show a clear similarity of trends in light response between our bioluminescence recordings of cultured whole brains exposed to the LP and anti-PER ICC analysis of whole brains of flies exposed to the LP in vivo, supporting previous conclusions that cultured whole brains of XLG-Per-Luc flies are excellent models for studying dynamic changes in the synchrony of PER activity induced by environmental cues such as light and temperature [24, 37].
Our bioluminescence measurements of synchrony in DD agree with our ICC measures of PER levels and previous studies which show an apparent progressive loss of synchrony and amplitude throughout most of the circuit over time [38, 39]. From their previously described role as core oscillators [27, 28, 36], the s-LNvs exhibit relatively robust rhythmic amplitude and synchrony in DD. The strong l-LNv amplitude and measurable phase coherence we observed even 2 days and beyond in DD is somewhat surprising based on expectations from earlier ICC studies [27–30] and our own ICC findings of l-LNv dampening of PER levels after 2 days (Figures 4–5). This is possibly due to (1) the improved temporal resolution of our longitudinal XLG-Per-Luc imaging approach, (2) the l-LNv loss of connection with the removed optic lobes or (3) lack of modulation from peripheral tissues. However, we find the same trends in light response for l-LNvs in brain cultures exposed to a LP ex vivo and l-LNv in the intact brains of adult flies exposed to a LP in vivo. This suggests that the l-LNv oscillators’ PER activity and their circuit connections are sufficiently intact in brain culture explants, though some light input and peripheral feedback information is obviously lost for cultured brains.
One of our most notable findings is that a phase advancing light pulse induces transient damping of the synchrony and rhythmicity of single neuron oscillators followed by the gradual emergence of a new state of strengthened synchrony that reproducibly varies across the circuit network. We call this dynamic process phase retuning. The new state of circuit synchrony is characterized by a light-induced phase shift that coincides with neurons exhibiting stronger rhythms that are better synchronized both within and across neuronal subgroups relative to DD. While we have not yet measured a comprehensive phase response curve, we expect that they will vary in a systematic fashion similar to behavioral phase response curves. Desynchrony may appear to be a negative consequence of the light pulse. However, recent work suggests that transient ‘phase tumbling’ [25] of the light entrainment process may be exploited for more rapid recovery from jetlag [25]. While much work has shown the importance of VIP peptidergic signaling in the SCN for maintaining robust rhythms [40–42], pharmacological treatment with VIP, GABAergic and vasopressin agents can transiently weaken oscillator function followed by more rapid entrainment [25, 43–45]. Temporarily weakening oscillator coupling and dephasing of rhythms appears to permit circuits to more easily reset to phase shifts and overly robust oscillator networks block entrainment [25, 43, 44, 46–49].
Previous work shows that circuit connectivity [29] organizes circadian behavior and electrical outputs of cell autonomous oscillators [50]. The Drosophila circadian circuit light initial response of desynchrony followed by phase retuning to a new circuit-wide synchrony pattern remarkably recapitulates many of the features we observe when LNvs are electrically hyperexcited [18, 51], suggesting that such responses are dictated by circuit properties. The relatively tight homogeneous light response that we measure in longitudinally imaged XLG-Per-Luc fly brains in the LNds is interesting as only half of the LNds express CRY [10]. This suggests a non-cell autonomous functional role for the LNds in light-induced circuit phase shift and maintaining behavior rhythmicity following exposure to a short light pulse. The LNds are the first neuronal subgroup to exhibit a rapid and coherent phase advance immediately following the light pulse. As suggested in [46], the LNds may first reset their own circadian oscillations before influencing other neuronal subgroups to reset and resynchronize their own molecular pacemakers. We propose that the LNds are the actual mediators of the whole circuit phase advance and that transient phase desynchrony in other neuronal subgroups enables them to be phase retuned which ultimately drives a light induced shift in the phase of behavioral rhythms. Sub-regions of the SCN also vary in oscillator response to light input and show a wave-like spatiotemporal pattern [52–54]. Comparisons of dissociated SCN cellular oscillators versus intact SCN slices suggest that many of the features of oscillator coordination are determined by anatomical connectivity [53, 55–58].
In summary, we show by whole circuit bioluminescence imaging of single circadian neurons and immunocytochemical analysis of PER activity in response to in vivo light exposure that a phase-advancing light pulse induces a circuit-wide spatiotemporal pattern of acute oscillator desynchrony followed by phase retuning to synchrony that varies across circadian neuronal subgroups. The general time course of this complex circuit-wide response imaged in whole brain explants closely matches that for behavioral entrainment in intact animals [12]. Based on the many organizational similarities of circadian circuits across the animal kingdom, entrainment appears to be constrained by connectivity of the circadian network. Our results support the hypothesis that temporarily weakened subsets of oscillators and their acute desynchrony are key initial features of entrainment. Broad features of this pattern of circadian circuit response to light may be generalized to humans and other mammals.
Experimental Procedures
A detailed description of reagents and protocols including organotypic whole brain culturing, bioluminescence imaging, immunocytochemical analysis of in vivo light response, and custom MATLAB scripts for quantitative analysis of PER activity can be found in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Drosophila whole brain explants cultured 6 days show circuit-wide response to light
Light induces transient loss of synchrony and individual oscillator rhythmicity
New state of phase retuned network synchrony gradually emerges after a light pulse
Neuronal subgroups exhibit distinct kinetic signatures of light response in vivo
Acknowledgments
We thank Ralf Stanewsky for sharing the XLG-Per-Luc transgenic fly line and polyclonal anti-PER antibody and Justin Blau for providing the monoclonal anti-PDF C7 antibody to the research community through the Developmental Studies Hybridoma Bank. We thank Keri Fogle, Eri Morioka, Jennifer Evans, Steven DeGroot and Sheeba Vasu for help with pilot experiments and technical advice; Vinh Nguy for assistance with bioluminescence time series analysis; Daniel Roberts for assistance with editing figures and movies; and Xiangmin Xu and Yulin Shi for advice on running MATLAB scripts. We thank the Optical Biology Core Facility at UC Irvine for the use of the LSM 700 confocal microscope and the Volocity software (PerkinElmer). This work was funded by NIH grants NS046750, NS078434, GM102965, and GM107405 and NSF IBN-0323466 to TCH and an NSF Graduate Research Fellowship DGE-1321846 to LR. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect those of the funding agencies.
Footnotes
Author Contributions: T.C.H and L.R. designed the experiments. L.R., A.M.G., J.H.H. and T.N. conducted the experiments. L.R., T.L.L. and T.C.H. analyzed the data. T.C.H., L.R., D.K.W., T.N. and T.L.L. wrote the manuscript.
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol. 1976;106:223–252. [Google Scholar]
- 2.Tauber E, Kyriacou BP. Insect photoperiodism and circadian clocks: models and mechanisms. J Biol Rhythms. 2001;16:381–390. doi: 10.1177/074873001129002088. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko M, Helfrich-Förster C, Hall JC. Spatial and Temporal Expression of the period andtimeless Genes in the Developing Nervous System of Drosophila: Newly Identified Pacemaker Candidates and Novel Features of Clock Gene Product Cycling. J Neurosci. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Ann Rev Physiol. 2010;72:551. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 7.Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaap J, Pennartz CM, Meijer JH. Electrophysiology of the Circadian Pacemaker in Mammals. Chronobiol Intl. 2003;20:171–188. doi: 10.1081/cbi-120019311. [DOI] [PubMed] [Google Scholar]
- 9.Ayaz D, Leyssen M, Koch M, Yan J, Srahna M, Sheeba V, Fogle KJ, Holmes TC, Hassan BA. Axonal injury and regeneration in the adult brain of Drosophila. J Neurosci. 2008;28:6010–6021. doi: 10.1523/JNEUROSCI.0101-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 11.Benito J, Houl JH, Roman GW, Hardin PE. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms. 2008;23:296–307. doi: 10.1177/0748730408318588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 13.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshii T, Ahmad M, Helfrich-Förster C. Cryptochrome mediates light-dependent magnetosensitivity of Drosophila’s circadian clock. PLoS Biol. 2009;7:e1000086. doi: 10.1371/journal.pbio.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sellix MT, Currie J, Menaker M, Wijnen H. Fluorescence/luminescence circadian imaging of complex tissues at single-cell resolution. J Biol Rhythms. 2010;25:228–232. doi: 10.1177/0748730410368016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen AL, Leise TL, Welsh DK. Bayesian statistical analysis of circadian oscillations in fibroblasts. J Theor Bio. 2012;314:182–191. doi: 10.1016/j.jtbi.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leise TL, Wang CW, Gitis PJ, Welsh DK. Persistent cell-autonomous circadian oscillations in fibroblasts revealed by six-week single-cell imaging of PER2:: LUC bioluminescence. PloS One. 2012;7:e33334. doi: 10.1371/journal.pone.0033334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheeba V. The Drosophila melanogaster circadian pacemaker circuit. J Genetics. 2008;8:485–493. doi: 10.1007/s12041-008-0071-x. [DOI] [PubMed] [Google Scholar]
- 19.Hamasaka Y, Rieger D, Parmentier ML, Grau Y, Helfrich-Förster C, Nässel DR. Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J Comp Neurol. 2007;505:32–45. doi: 10.1002/cne.21471. [DOI] [PubMed] [Google Scholar]
- 20.Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, Nässel DR. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. DN1p Circadian Neurons Coordinate Acute Light and PDF Inputs to Produce Robust Daily Behavior in Drosophila. Curr Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and Temperature Control the Contribution of Specific DN1 Neurons to Drosophila Circadian Behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins B, Kane EA, Reeves DC, Akabas MH, Blau J. Balance of Activity between LNvs and Glutamatergic Dorsal Clock Neurons Promotes Robust Circadian Rhythms in Drosophila. Neuron. 2012;74:706–718. doi: 10.1016/j.neuron.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veleri S, Brandes C, Helfrich-Förster C, Hall JC, Stanewsky R. A Self-Sustaining, Light-Entrainable Circadian Oscillator in the Drosophila Brain. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 25.An S, Harang R, Meeker K, Granados-Fuentes D, Tsai CA, Mazuski C, Kim J, Doyle FJ, Petzold LR, Herzog ED. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc Natl Acad Sci USA. 2013;110:E4355–E4361. doi: 10.1073/pnas.1307088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonze D, Bernard S, Waltermann C, Kramer A, Herzel H. Spontaneous synchronization of coupled circadian oscillators. Biophys J. 2005;89:120–129. doi: 10.1529/biophysj.104.058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Sehgal A. Role of Molecular Oscillations in Generating Behavioral Rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 28.Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Bio. 2003;1:e13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klarsfeld A, Malpel S, Michard-Vanhée C, Picot M, Chélot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheeba V, Sharma VK, Gu H, Chou YT, O’Dowd DK, Holmes TC. Pigment dispersing factor-dependent and-independent circadian locomotor behavioral rhythms. J Neurosci. 2008;28:217–227. doi: 10.1523/JNEUROSCI.4087-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large Ventral Lateral Neurons Modulate Arousal and Sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian-and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophys. 2007;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang YH, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dissel S, Hansen Celia N, Özkaya Ö, Hemsley M, Kyriacou Charalambos P, Rosato E. The Logic of Circadian Organization in Drosophila. Curr Biol. 2014;24:2257–2266. doi: 10.1016/j.cub.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helfrich-Förster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microscopy Res Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- 37.Sehadova H, Glaser FT, Gentile C, Simoni A, Giesecke A, Albert JT, Stanewsky R. Temperature Entrainment of Drosophila’s Circadian Clock Involves the Gene nocte and Signaling from Peripheral Sensory Tissues to the Brain. Neuron. 2009;64:251–266. doi: 10.1016/j.neuron.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf Neuropeptide Gene Mutation and Ablation of PDF Neurons Each Cause Severe Abnormalities of Behavioral Circadian Rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC2 Receptor Is Essential for Circadian Function in the Mouse Suprachiasmatic Nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 41.Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP-and PHI-deficient mice. Amer J Physiol Reg Int Comp Physiol. 2003;285:R939–R949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- 42.Brown T, Colwell CS, Waschek J, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophys. 2007;97:2553–2558. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freeman GM, Jr, Krock RM, Aton SJ, Thaben P, Herzog ED. GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron. 2013;78:799–806. doi: 10.1016/j.neuron.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, Dong X, Tsujimoto G, Okuno Y, Doi M, Okamura H. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science. 2013;342:85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- 45.Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80:973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamba P, Bilodeau-Wentworth D, Emery P, Zhang Y. Morning and Evening Oscillators Cooperate to Reset Circadian Behavior in Response to Light Input. Cell Rep. 2014;7:601–608. doi: 10.1016/j.celrep.2014.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatori M, Gill S, Mure LS, Goulding M, O’Leary DD, Panda S. Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife. 2014;3:e03357. doi: 10.7554/eLife.03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buhr E, Van Gelder RN. The making of the master clock. eLife. 2014;3:e04014. doi: 10.7554/eLife.04014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb AB, Taylor SR, Thoroughman KA, Doyle FJ, III, Herzog ED. Weakly circadian cells improve resynchrony. PLoS Comp Biol. 2012;8:e1002787. doi: 10.1371/journal.pcbi.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nitabach MN, Blau J, Holmes TC. Electrical Silencing of Drosophila Pacemaker Neurons Stops the Free-Running Circadian Clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 51.Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, Shinohara K. Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J Neurosci Res. 2005;82:622–630. doi: 10.1002/jnr.20677. [DOI] [PubMed] [Google Scholar]
- 53.Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One. 2011;6:e15869. doi: 10.1371/journal.pone.0015869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foley NC, Tong TY, Foley D, LeSauter J, Welsh DK, Silver R. Characterization of orderly spatiotemporal patterns of clock gene activation in mammalian suprachiasmatic nucleus. Eur J Neurosci. 2011;33:1851–1865. doi: 10.1111/j.1460-9568.2011.07682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 56.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans JA, Pan H, Liu AC, Welsh DK. Cry1/circadian rhythmicity depends on SCN intercellular coupling. J Biol Rhythms. 2012;27:443–452. doi: 10.1177/0748730412461246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noguchi T, Wang LL, Welsh DK. Fibroblast PER2 circadian rhythmicity depends on cell density. J Biol Rhythms. 2013;28:183–192. doi: 10.1177/0748730413487494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leise TL, Harrington ME. Wavelet-based time series analysis of circadian rhythms. J Biol Rhythms. 2011;26:454–463. doi: 10.1177/0748730411416330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.