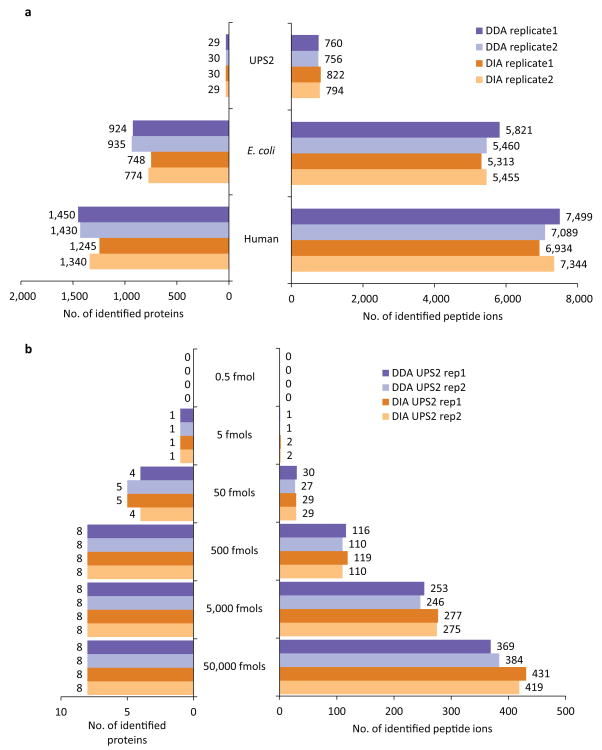
Fig. 3. Untargeted peptide and protein identification using DDA and DIA data from UPS2, E. coli, and human cell lysate samples.
(a) The number of peptide ions and proteins identified by X! Tandem search engine at 1% FDR in DDA and in DIA (SWATH) data from UPS2, E. coli, and human cell lysate samples. (b) The number of peptide ions and protein identifications (X! Tandem) in each replicate of the UPS2 sample DDA and DIA data plotted separately for proteins of different abundance (in UPS2 samples 48 proteins span 5 orders of magnitude of abundance ranging from 0.5 to 50,000 fmoles with 8 proteins in each abundance range).