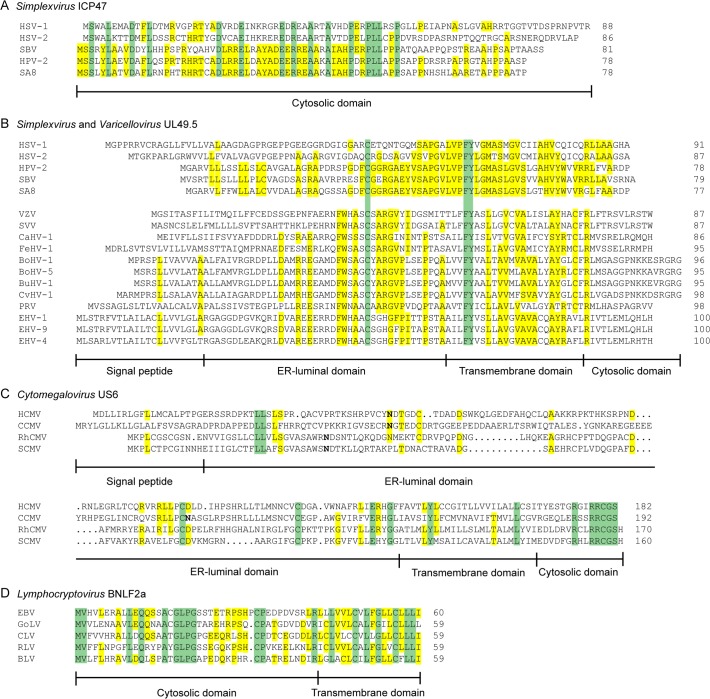
Fig 2. Alignments of the amino acid sequences of selected herpesvirus-encoded TAP-inhibitors.
A) simplexvirus ICP47 orthologs, B) simplexvirus (upper 5 lines) and varicellovirus (lower 12 lines) UL49.5 orthologs, C) cytomegalovirus US6 orthologs, and D) lymphocryptovirus BNLF2a orthologs. The alignments of predicted primary translation products were made using ClustalW, followed by manual adjustment. The number of residues in each sequence is shown on the right. Green highlights residues that are conserved in all sequences, and yellow highlights residues that are conserved in a majority. Bold N residues in US6 indicate potential N-linked glycosylation sites. An illustrationof sequence disposition is shown below each alignment, with approximate boundaries displayed.