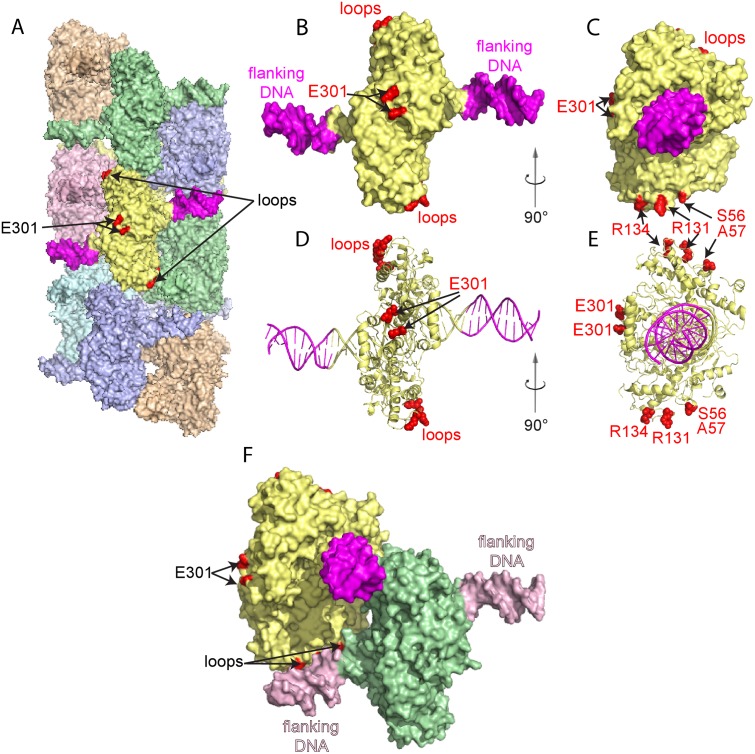
Fig 3. Location of additional flanking DNA and single site substitutions in SgrAI.
A. Space filling representation of the model of the SgrAI/DNA run on oligomer derived from cryo-EM single particle reconstruction[10]. Each DNA bound SgrAI dimer is colored differently, and the outer 11 bp of flanking DNA and sites of point mutations are shown in magenta and red, respectively, in the central DNA bound SgrAI dimer [10]. E301 is located on the outer surface of SgrAI distal from the DNA binding site. B. Space filling model of a single DNA bound SgrAI dimer with the outer 11 bp of DNA flanking the SgrAI recognition sequence colored magenta and sites of single site substitutions (E301 and loops containing S56, S57, R131 and R134) highlighted in red. C. As in B, rotated 90°. D and E. Ribbon diagram of B and C, respectively. F. Close-up view of the interactions between loop residues 56–60 and 127–134 of one SgrAI chain (yellow) and the flanking DNA (light pink) bound to a neighboring SgrAI (green) in the run-on oligomer.