FIGURE 3.
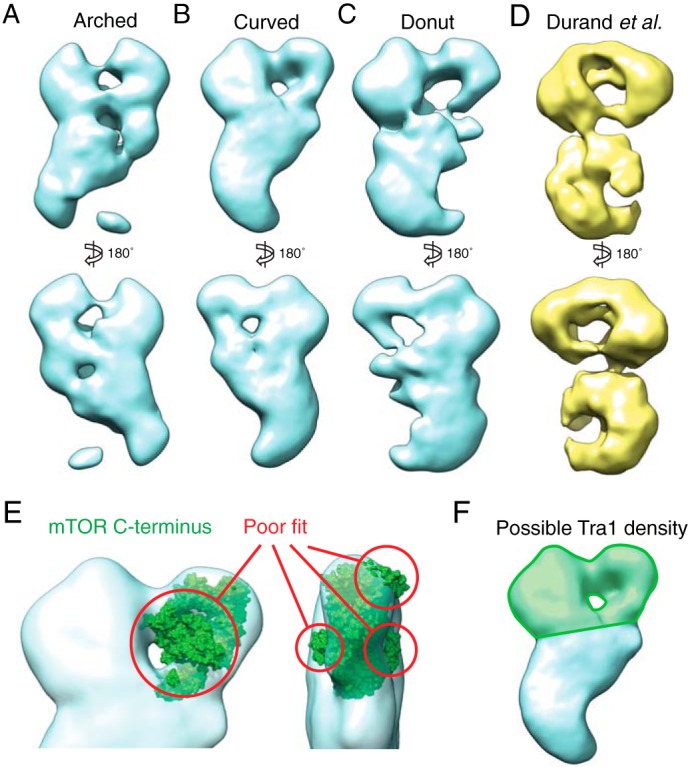
Three-dimensional reconstructions of the three SAGA conformations. A–C, models were generated de novo from both tilted and untilted particles that correspond to each SAGA conformation using the random conical tilt method. D, three-dimensional reconstruction of SAGA purified without gradient fixation by Durand et al. (Electron Microscopy Data Bank code 2693). E, the mTOR C-terminal crystal structure (Protein Data Bank code 4JSV), which shares a high degree of secondary structure similarity with the Tra1 C terminus, does not fit within the region of the head of SAGA previously proposed to contain Tra1. F, the region that likely contains the entire Tra1 structure is highlighted in green.
