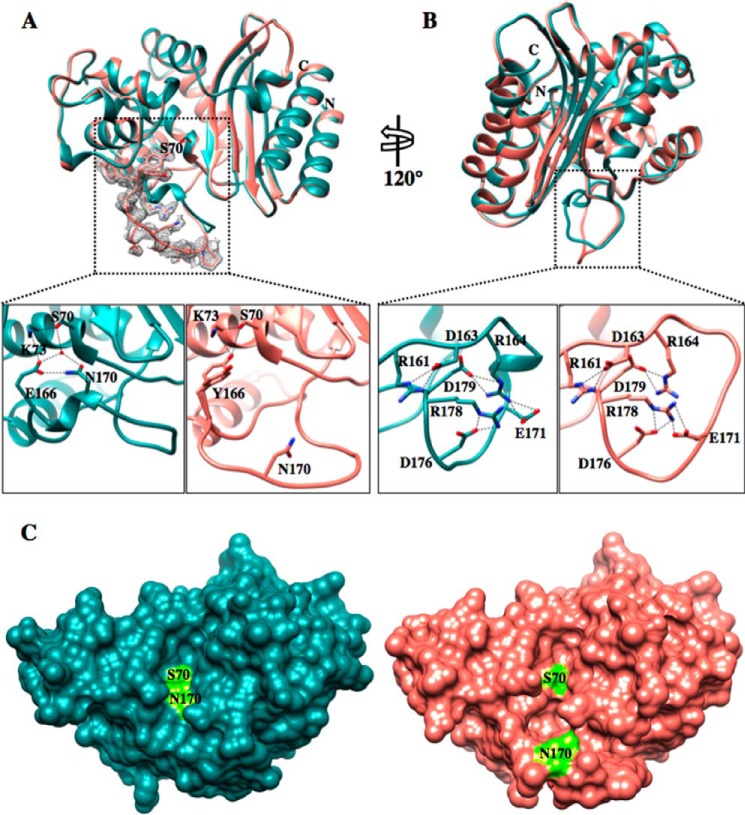
FIGURE 3.
Crystal structure of the TEM-1 W165Y/E166Y/P167G/L201P mutant. A, top panel, TEM-1 in dark cyan (PDB code 1XPB, 1.9 Å resolution) aligned with the TEM-1 W165Y/E166Y/P167G/L201P (salmon) enzyme. The active-site Ser-70 is represented in both structures in stick model. The simulated annealing omit difference map contoured at ∼3 σ for residues 164–174 in the W165Y/E166Y/P167G/L201P structure is shown as a gray mesh and reveals the two different conformations that the Tyr-166 adopts in the W165Y/E166Y/P167G/L201P mutant structure. Bottom panels, detailed view of the catalytic apparatus of TEM-1 (left bottom panel) and proposed apparatus of W165Y/E166Y/P167G/L201P mutant (right bottom panel). In the mutant, the catalytic water molecule is not present because of the bulkier tyrosine residue and the movement of Asn-170. B, top panel, 120° rotation of the structure alignment with the dotted black box indicating the Ω-loop. Bottom panels, view of the Ω-loop electrostatic network. The Ω-loop structure of the mutant is maintained by the preserved salt bridges among the charged residues within the Ω-loop. C, surface representation of TEM-1 (dark cyan) (PDB code 1XPB) and W165Y/E166Y/P167G/L201P (salmon) structures. The active site Ser-70 and Asn-170 are labeled and represented in fluorescent green. The active site cavity of the mutant is enlarged and elongated, forming an L shape. In contrast, the TEM-1 active site is shallow and narrow.