FIGURE 5.
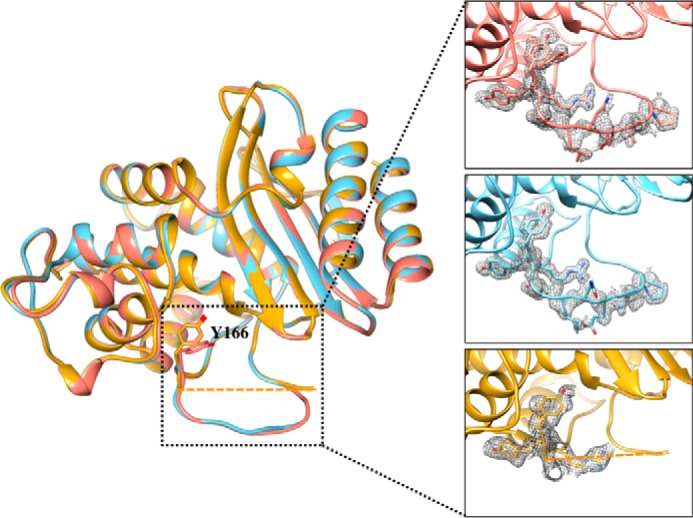
Alignment of the three structures of the TEM-1 W165Y/E166Y/P167G mutants solved in this study. In salmon is the structure of W165Y/E166Y/P167G/L201P. The structure of W165Y/E166Y/P167G/M182T is shown in gold. Residues 168–174 of the Ω-loop of W165Y/E166Y/P167G/M182T showed very little density, and were not modeled in the final structure (residues 167 and 175 are connected with a dashed line). The structure of S70G/W165Y/E166Y/P167G is shown in light blue. The Tyr-166 residue assumes the same conformation in the three structures; however, in the L201P structure, an addition conformation of Tyr-166 is observed. The inset shows a detailed view of the Ω-loop in the three structures, and the simulated annealing omit difference maps contoured at ∼3 σ are shown as a gray mesh for residues 164–174 in the W165Y/E166Y/P167G/L201P and S70G/W165Y/E166Y/P167G structures and for residues 164–167 in the W165Y/E166Y/P167G/M182T structure.
