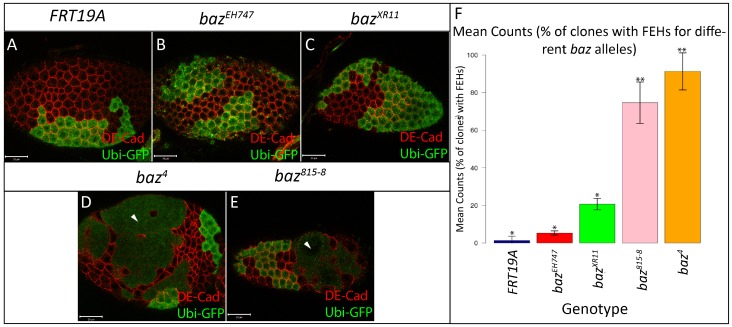
Fig. 1. Different baz alleles display different FE phenotypes.
FE clones marked by absence of GFP were generated using the directed mosaic system by expressing UAS-Flp under the control of e22c-Gal4. FRT19A (A), bazEH747 (B) and bazXR11 (C) mutant FE cells maintain a continuous epithelium while baz4 (D) and baz815-8 (E) mutant cells fail to maintain a continuous epithelium resulting in large holes in the FE, exposing the underlying nurse cells (white arrowheads). (F) Bar graph presenting quantitative analysis of the FEH phenotype. The number of baz4, baz815-8, bazEH747, bazXR11 and FRT19A mosaic follicles with holes in the lateral epithelium were counted and presented as an average percentage from three repetitions of n = 50 (88% in baz4, 75% in baz815-8, 21% in bazXR11, 5% in bazEH747, 1% in FRT19A). At p<0.001, genotypes with same number of stars are not significantly different while those with different number of stars are significantly different. Error bars represent s.d. Scale bars = 20 µm.