Figure 3.
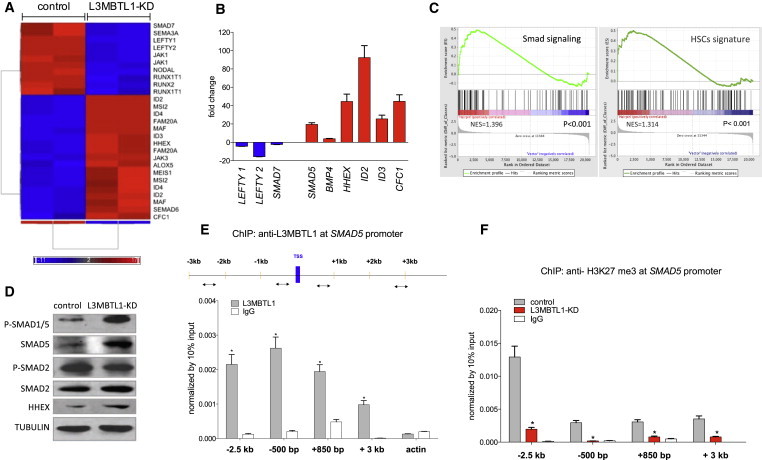
L3MBTL1 Transcriptionally Represses SMAD5
(A) GEP of undifferentiated L3MBTL1-KD iPSCs, compared to controls, based on microarray analysis. We utilized independent clones of iPSCs (generated from cord blood CD34+ cells), which we independently infected with lentiviral vectors expressing shRNAs against L3MBTL1.
(B) mRNA expression levels of several upregulated and downregulated genes, identified by microarray analysis, was confirmed by qPCR. Data were normalized by GAPDH expression and shown as shRNA versus control. The data represent the mean ± SD of the three independent experiments.
(C) GSEA for the combined set of SMAD and hematopoietic stem cells genes compared with the differentially expressed genes in L3MBTL1-KD iPSCs.
(D) Expression levels of phospho SMAD1/5, total SMAD5, phospho SMAD2, total SMAD2, and HHEX were evaluated in L3MBTL1-KD and control iPSCs by western blot. Tubulin served as the loading control.
(E) iPSCs were crosslinked with 1% formaldehyde and immunoprecipitated with anti-L3MBTL1 or IgG antibody (as a nonspecific control). Plotted values are relative enrichments (y axis) to 10% input and measured for sites in the SMAD5 promoter and actin (x axis). The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test.
(F) L3MBTL1-KD and control iPSCs were crosslinked with 1% formaldehyde and immunoprecipitated with an anti-H3K27 tri-methyl antibody or an IgG antibody (as a nonspecific control). Plotted values are relative enrichments (y axis) to 10% input and measured for sites in the SMAD5 promoter (x axis). The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test. See also Figure S3.
