Figure 2.
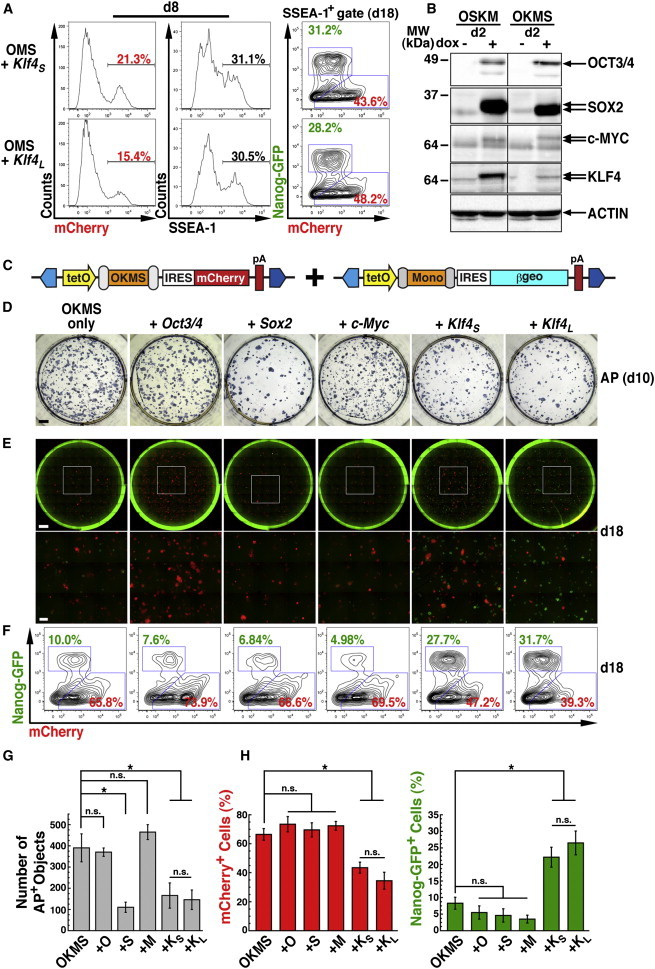
KLF4 Stoichiometry Affects Reprogramming Phenotypes
(A) Reprogramming with polycistronic OMS + monocistronic Klf4S or Klf4L leads to nearly identical phenotypes, as analyzed by FACS for mCherry or SSEA-1 on d8 (left) and mCherry versus Nanog-GFP on d18 (right).
(B) Western blot analysis of OCT3/4, SOX2, c-MYC, and KLF4 in OSKM or OKMS transfected MEFs cultured for 2 days with or without dox treatment. Actin was used as a loading control. Arrows show the two KLF4 isoforms. Note that SOX2 and c-MYC also differ in size based on their positions relative to 2A peptides in the polycistronic cassette (Figure 1D). Uncropped data are provided in Figure S2G.
(C–F) Supplementation of PB-TAC-OKMS was performed by co-transfection of additional factors in PB-TAB transposons. β-geo, lacZ-neo fusion gene.
(D) AP staining on d10 after transfection with OKMS alone or in combination with Oct3/4, Sox2, c-Myc, Klf4S, or Klf4L. Scale bar, 4,000 μm.
(E) Day 18 fluorescence microscopy of entire wells (composite 10 × 10 fields) or selected insets (3 × 3 subfields) for mCherry+ and Nanog-GFP+. Scale bars, 4,000 μm (full well) and 1,000 μm (inset).
(F) FACS analysis of mCherry and Nanog-GFP fractions in the d18 SSEA-1+ population of the cultures shown in (E). The results in (D)–(F) are representative of the results of at least three independent experiments (summarized in G and H).
(G) Quantification of the effects of factor supplementation on AP+ colony formation (D). Means ± SE for three independent experiments. n.s., not significant. ∗p < 0.05, Student’s t test.
(H) Percentages of mCherry+ and Nanog-GFP+ populations from FACS analysis of factor supplementation (F). Means ± SE for three independent experiments. n.s., not significant. ∗p < 0.05, Student’s t test.
See also Figure S2.
