Figure 3.
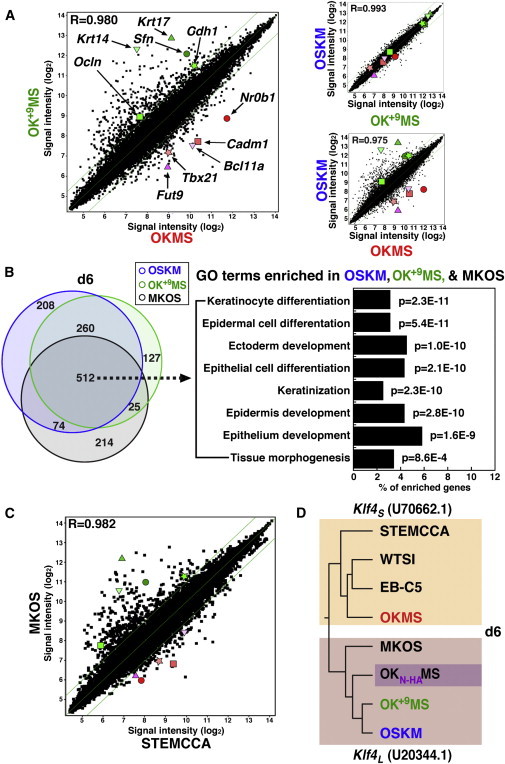
Gene Expression Patterns Reveal Klf4 Variants
(A) Pairwise comparison of global gene expression in mCherry-positive cells on d6 from OSKM, OKMS, and OK+9MS reprogramming. Diagonal lines indicate 2-fold changes in log2 signal intensity. Genes chosen for indexing are highlighted. Green: Krt17, triangle; Sfn, circle; Ocln, square; Cdh1, star; Krt14, inverted triangle. Red: Fut9, triangle; Nr0b1, circle; Cadm1, square; Tbx21, star; Bcl11a, inverted triangle.
(B) Left: Venn diagram showing overlapping gene activation versus lacZ-MEFs in the Klf4L cluster. Right: GO analysis of 512 shared d6 genes, arranged in order of p value and indicating the proportion of genes represented for each enriched GO term. Cutoff p = 1.0E-3.
(C) Pairwise comparison of global gene expression in mCherry-positive cells on d6 from MKOS and STEMCCA reprogramming. Highlighted genes are those indicated in (A).
(D) Dendogram resulting from unsupervised clustering of OSKM, OKMS, OK+9MS, OKN-HAMS, MKOS, STEMCCA, EB-C5, and WTSI mCherry d6 array samples. Segregation of the vectors based on the nature of the KLF4 N terminus is indicated.
