Abstract
Red blood cell (RBC) folate levels are established at the time of erythropoiesis and therefore provide a surrogate biomarker for the average folate status of an individual over the preceding four months. Folates are present as folylpolyglutamates, highly polar molecules that cannot be secreted from the RBCs, and must be converted into their monoglutamate forms prior to analysis. This was accomplished using an individual’s plasma pteroylpolyglutamate hydrolase by lysing the RBCs in whole blood at pH 5 in the presence of ascorbic acid. Quantitative conversion of formylated tetrahydrofolate derivatives into the stable 5,10-methenyltetrahydrofolate (5,10-MTHF) form was conducted at pH 1.5 in the presence of [13C5]-5-formyltetrahydrofolate. The resulting [13C5]-5,10-MTHF was then used as an internal standard for the formylated forms of tetrahydrofolate that had been converted into 5,10-MTHF as well any 5,10-MTHF that had been present in the original sample. A stable isotope dilution liquid chromatography-multiple reaction monitoring/mass spectrometry method was validated and then used for the accurate and precise quantification of RBC folic acid, 5-methyltetrahydrofolate (5-MTHF), tetrahydrofolate (THF), and 5,10-MTHF. The method was sensitive and robust and was used to assess the relationship between different methylenetetrahydrofolate reductase (MTHFR) 677C>T genotypes and RBC folate phenotypes. Four distinct RBC folate phenotypes could be identified. These were classified according to the relative amounts of individual RBC folates as type I (5-MTHF >95%; THF <5%; 5,10-MTHF <5%), type II (5-MTHF <95%; THF 5% to 20%; 5,10-MTHF <5%), type III (5-MTHF >55%; THF >20%; 5,10-MTHF >5%), and type IV (5-MTHF <55%; THF >20%; 5,10-MTHF >5%).
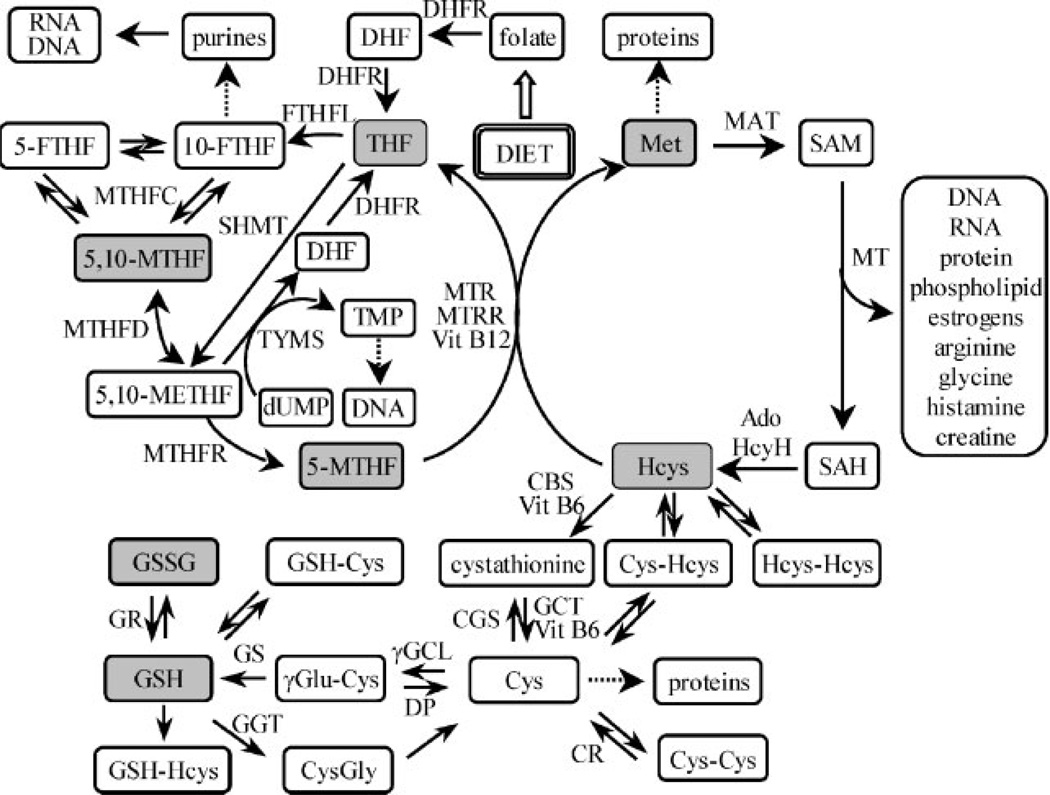
Folate/homocysteine metabolism provides one-carbon units for many essential biological processes.1–4 In particular, it enables the methylation of substrates including DNA, proteins and lipids, and the generation of thymidylate and purines, important functions that require different intracellular folate derivatives (Fig. 1).5 Low folate status per se is associated with elevated levels of circulating homocysteine (hyperhomocysteinemia),4 and a phenotype characterized by low red blood cell (RBC) and serum folate together with high homocysteine has been implicated in many diverse human pathologies ranging from neural tube defects such as spina bifida6,7 to aging-related conditions such as cardiovascular disease3 and colorectal cancer.8,9 Whether one or both of these two biochemical variables are causally involved in pathogenic change or are markers of the direct etiologic agent remains a matter of debate. However, there is clearly a potential for folate dysregulation to negatively impact several key cellular functions. In addition, the potentially deleterious effects of hyperhomocysteinemia are a consequence of inadequate levels of the methyl donor 5-methyltetrahydrofolate (5-MTHF) (Fig. 1).5,10 Folate/homocysteine metabolism also modulates glutathione biosynthesis through the cystathionine/cysteine pathway, which is in turn crucial for controlling intracellular redox status (Fig. 1).11 This suggests that an understanding of the parameters which determine the intracellular distribution of folate derivatives would provide the foundation for detailed characterization of the downstream pathways supported by each individual derivative.
Figure 1.
Abbreviations: BHMT=betaine homocysteine methyltransferase; CBL=cystathionine β-lyase; CBS=cystathionine β-synthase; CGS=cystathionine γ-synthase; CR=cysteine reductase; Cys=cysteine; DHFR=dihydrofolate reductase; DP=dipeptidases; dUMP=2′-deoxyuridine monophosphate; FTHFL=formate tetrahydrofolate ligase; GGT=γ-glutamyltran-speptidase; GGCL=γ-glutamylcysteinyl ligase; GR=glutathione reductase; GS=glutathione synthase; GSH=glutathione; Hcys=homocysteine; MAT=methionine adenosyltransferase; MTHFC=methylenetetrahydrofolate cyclohydrolase; MTHMD=methylenetetrahydrofolate dehy-drogenase; MTHFR=methylenetetrahydrofolate reductase; MTR=methionine synthase; MTRR=methionine synthase reductase; SAH=S-adenosylhomocysteine; SAM=S-adenosylmethionine; SHMT=serine hydroxymethyltransferase; TMP=thymidine monophosphate; TYMS=thymidylate synthase; Vit=vitamin.
Over the past ten years several functional polymorphisms in enzymes involved in folate/homocysteine metabolism have been described.12 The functional polymorphism with the most readily observed impact on phenotype is the C to T transition at nucleotide 677 (677C>T) of the methylenetetrahydrofolate reductase (MTHFR) gene, which results in a change in amino acid residue from Ala>Val at position 222, located at the bottom of the (βα)8 barrel of the catalytic domain of the enzyme.13 The 677T allele encodes an enzyme that is ‘thermolabile’ and less efficient at generating the 5-MTHF that is needed for both homocysteine remethylation and the generation of S-adenosyl methionine for methylation reactions (Fig. 1). It is well established that MTHFR 677TT homozygotes with low folate status are at greatly increased risk of being hyperhomocysteinemic.14 Selhub and colleagues have established that in the RBCs of individuals with this genotype, 5-MTHF comprises only 60% of total RBC folate, whereas this form predominates in the RBCs of their 677CC peers.15,16 Subsequently, Smulders et al. showed that the MTHFR C677T genotype is the primary determinant of non-methylfolate accumulation in RBCs.17 The homozygous MTHFR 677TT genotype confers a significantly increased risk of many of the conditions with which a low folate, high homocysteine phenotype has been associated, for example, approximately 2-fold for spina bifida,18 and 1.15-fold for cardiovascular disease.19 However, the excess individual risk of developing such conditions in relation to their prevalence is insufficient to warrant genetic testing and counseling. Therefore, there is a need to establish laboratory methods to define the degree of variation in the ‘folate phenotypes’ between and within the three MTHFR 677C>T genotypes in order to determine whether there are subsets of TT homozygotes, and possibly of CT heterozygotes, with extreme phenotypes that may be associated with greatly enhanced risk. Individuals falling into such subsets might benefit from early identification and intervention.
There is a rich literature describing methodology that can be employed for folate analysis,20 including microbiological,21,22 competitive binding assays (radioassays),22,23 liquid chromatography (LC)/electrochemical detection,24,25 LC/fluoroescence,26 gas chromatography/mass spectrometry (GC/MS),27,28 and LC-multiple reaction monitoring (MRM)/MS.17,29–39 Unfortunately, many of the available methods have limited utility for rigorous population studies because of the ease with which RBC folates can degrade and/or interconvert during the analytical procedure. This makes it preferable to utilize methodology based on stable isotope dilution LC-MRM/MS, which can efficiently correct for such problems.26,27,39 Specificity is conferred by requirements that the folates must have the same LC retention time as their corresponding [13C5]-labeled internal standards, as well as the same precursor ion and the same product ion as authentic unlabeled standards. No other technique can provide this level of specificity. Although this approach has been described in a number of recent publications,29–39 no method has been validated for quantification of all the key RBC folates in human subjects with well-defined genotypes. We report the development of a validated stable isotope dilution LC-MRM/MS method for the analysis of RBC and plasma folates and its use to identify different phenotypes between and within the MTHFR 677C>T homozygous CC, heterozygous CT, and homozygous TT genotypes.
EXPERIMENTAL
Materials
Supelco LC-18 3mL solid-phase extraction (SPE) cartridges were obtained from Supelco (Bellefonte, PA, USA). HPLC grade water, methanol, and acetonitrile were obtained from Fisher Scientific Co. (Fair Lawn, NJ, USA). 2-Mercaptoethanol was obtained from Bio-Rad Laboratories (Hercules, CA, USA). Folic acid (FA), THF, 5-MTHF, acetic acid, and ascorbic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). 5,10-MTHF, 5,10-methylenetetrahydrofolic acid (METHF), 5-formyltetrahydrofolic acid (5-FTHF), 10-formyltetrahydrofolic acid (10-FTHF) and pteroylhepta-γ-L-glutamic acid were obtained from Schircks Laboratories (Jona, Switzerland). [13C5]-FA, [13C5]-THF, [13C5]-5-MTHF, and [13C5]-5-FTHF were obtained from Eprova AG (Schaffhausen, Switzerland). Argon and liquid nitrogen were obtained from BOC Gases (Bellmawr, NJ, USA).
Samples from individuals with defined MTHFR 677C>T genotypes
Blood samples were obtained from female subjects enrolled in two ongoing studies of folate and homocysteine metabolism at the University of Pennsylvania School of Medicine. Major exclusionary criteria for the studies were use of anti-folate medications and pregnancy. Both studies were approved by the Institutional Review Board of the University of Pennsylvania School of Medicine, and all subjects gave written informed consent. The samples used were from the first five Caucasian subjects under the age of 50 years in each of the MTHFR 677C>T genotype classes (i.e., CC, CT, and TT) who had been recruited for each of the studies.
Whole blood samples
Blood samples for RBC folate analysis were drawn into 4mL EDTA (purple top) tubes and placed in the dark until processed. Each tube was gently inverted (without shaking or foaming contents) six times prior to transfer of 1 mL aliquots to separate 15 mL tubes and addition of 9 mL aqueous 1% ascorbic acid solution.20,40 Each tube was gently inverted six times, left in the dark at room temperature for 30 min, and then gently inverted six more times. Contents were transferred to dark amber 2 mL tubes, frozen on dry ice, and stored at −80°C until analyzed.
Plasma samples
Blood samples for plasma folate analysis were drawn into 4 mL EDTA (purple top) tubes and placed in the dark until processed. The tubes were centrifuged in a Eurotech (Beaconsfield, UK) Z-150 A centrifuge at 1100 g for 5 min. The resulting plasma was transferred to dark amber 2 mL tubes, frozen on dry ice and stored at −80°C until analyzed.
DNA isolation and MTHFR 677C>T genotyping
DNA was extracted from whole blood using the QIAamp@ DNA Mini Kit (Qiagen). MTHFR 677C>T (rs1801133) allelic discrimination was performed using a TaqMan 5′ Nuclease real-time polymerase chain reaction (PCR) assay on a DNA Engine Opticon 2 continuous fluorescence detection system (MJ Research, Waltham, MA, USA). PCR amplification was performed using 2 µL of sample DNA, 1× TaqMan Universal PCR MasterMix (Applied Biosystems, Foster City, CA, USA), 0.5µM of each primer (5′-GCAGGGAGCTTTGAGGCTGACC-3′ and 5′-TGGGGCAAGTGATGCCCATGT-3′), 50 nM ‘T’-specific probe (6FAM-ATGAAATCGACTCCCGC-MGBNFQ) and 100 nM ‘C’-specific probe (VIC-ATGAAATCGGCTCCCGC-MGBNFQ). Probe sequences were derived from the SNP500Cancer website.41 They were custom synthesized by Applied Biosystems (Foster City, CA, USA). PCR was performed with an initial incubation at 95°C for 10 min, followed by 60 cycles of denaturation at 92°C for 30s and extension/5′ nuclease step at 56°C for 1 min. Dual fluorescence was detected after each extension 5′ nuclease step. Genotype interpretations were performed using OpticonMonitor Analysis software version 2.02 (MJ Research, Ramsey, MN, USA).
Preparation of standard and QC solutions
All procedures were performed under conditions of decreased laboratory lighting. Standards and quality control (QC) solutions were prepared using certified volumetric flasks with certified Hamilton microsyringes. Stock solution I for 5-MTHF, 5-FTHF, and THF (100µg/mL) was prepared by dissolving 5-MTHF (1 mg, 2.2µmol), 5-FTHF (1 mg, 2.1µmol) and THF (1 mg, 2.2µmol) in 20 mM phosphate buffer (pH 7.2) containing cysteine (1mg/mL) in a 10 mL volumetric flask. Phosphate buffer was added to the mark, an aliquot (200 µL) of the solution was removed for ultraviolet (UV) spectrophotometry using a Beckman Du530 UV spectrophotometer (Beckman Instruments, Fullerton, CA, USA), and ascorbic acid (100 mg) was then added to the volumetric flask. Stock solution I for FA (100 µg/mL; 1 mg, 2.3 µmol) was prepared in a similar manner, except that 20mM phosphate buffer (pH 7.2) was used without the addition of cysteine or ascorbic acid, and an aliquot (200 µL) was removed for UV analysis. Concentrations of each folate were determined after a 20-fold dilution with phosphate buffer (5-MTHF λmax = 290 nm ε = 31.7 L/mmol/cm; 5-FTHF λmax = 285 nm ε = 37.2 L/mmol/cm; THF λmax = 297 nm ε = 29.1 L/mmol/cm; FA λmax = 282 nm ε = 27.6L/mmol/cm).42 Stock solution I for 5,10-MTHF (100 µg/mL; 1 mg, 2.2 µmol) was prepared in a similar manner except that it was dissolved in 5 mM hydrochloric acid. The concentration was confirmed using a 20-fold dilution with 0.01% acetic acid at pH 3 (5,10-MTHF λmax = 360 nm ε = 25.1 L/mmol/cm).42 All the standards and QC samples were prepared by serial dilutions from these stock solutions. Working solutions were prepared every 4 weeks and their concentrations were checked by UV spectrophotometry before use. Heavy isotope standard solutions were prepared in the same way as the relevant unlabeled folate standards.
Mass spectrometry
Mass spectrometry was conducted using an Applied Biosystems API4000 triple-quadrupole mass spectrometer (Foster City, CA, USA) equipped with a turboionspray source and operated in the positive ion mode. Operating conditions were as follows: source temperature, 450°C; spray voltage, 5.0 kV; collision cell exit potential, 10 V; collision gas pressure, 6 psi; curtain gas, 30psi; Gas1, 40psi; and Gas2, 30psi. LC-MRM/MS was conducted using the following transitions for FA, m/z 442 (MH+) to m/z 295 (MH+-γ-glutamate); [13C5]-FA, m/z 447 (MH+) to m/z 295 (MH+-γ-glutamate); THF, m/z 446 (MH+) to m/z 299 (MH+-γ-glutamate); [13C5]-THF, m/z 451 (MH+) to m/z 299 (MH+-γ-glutamate); 5-MTHF, m/z 460 (MH+) to m/z 313 (MH+-γ-glutamate); [13C5]-5-MTHF, m/z 465 (MH+) to m/z 313 (MH+-γ-glutamate); 5,10-MTHF, m/z 456 (M+) to m/z 412 (M+-CO2); [13C5]-5,10-MTHF m/z 461 (M+) to m/z 416 (M+-13CO2); 5-FTHF m/z 474 (MH+) to m/z 327 (MH+-γ-glutamate); [13C5]-5-FTHF m/z 479 (MH+) to m/z 327 (MH+-γ-glutamate). Collision offset energies for FA, THF, 5-MTHF, 5,10-MTHF, and 5-FTHF were 21,29,27,41, and 29 V, respectively.
Liquid chromatography
Chromatography was performed using an Agilent 1100 separation module (Palo Alto, CA, USA) equipped with a Leap autoinjector (CTC Analytics AG, Switzerland). Gradient elution of the folates was conducted in the linear mode using a YMC ODS-AQ column (150 × 2.0 mm i.d., 3µm, 120 Å Waters Inc., Milford, MA, USA). Mobile phase A was 1% acetic acid in water and mobile phase B was 1% acetic acid in methanol/acetonitrile (4:1). The flow rate was 200 µL/min. The gradient conditions were as follows: 0min, 1% B; 10min, 91% B; 13min, 91% B; 14min, 1% B, and 25min 1%B. The samples (200 µL) were maintained at 4°C in the autosampler tray, and injections of 50µL were made. The gradient was started immediately after the sample injection. The column effluent was diverted to waste for the first 8min of the analysis to prevent extraneous and endogenous materials from entering the mass spectrometer.
Whole blood sample preparation for RBC folate analysis
Eight calibration standards were prepared in the range 4.5 to 900 nmol/L in 1% ascorbic acid and 10mM 2-mercaptoethanol (to prevent oxidation). To 500 µL of whole blood (1:10 diluted with 1% ascorbic acid) was added 20 µL of internal standard solution (150 pg/µL each of [13C5]-FA, [13C5]-THF, [13C5]-5-MTHF, and [13C5]-5-FTHF). For hydrolysis of folylpolyglutamates, 1 N sodium hydroxide (6µL) was added to each tube to adjust the pH to 5.0 and the samples were mixed immediately. In selected experiments, pteroylhepta-γ-L-glutamic acid (5 ng) was added to replicate (n = 5) RBC preparations from five different subjects in order to monitor the efficiency of folylpolyglutamate hydrolysis. Samples were degassed with argon, which was also used to flush the tubes. The covered samples were kept at room temperature for 4h in the dark in order to complete the hydrolysis of the polyglutamated forms of the folates. Before further purification using SPE columns, 1 mL water containing 1% ascorbic acid and 1% methanol was added.
Sample preparation for analysis of plasma folates
To each plasma sample (300 µL) was added water (200 µL) containing 1% ascorbic acid and 10 mM 2-mercaptoethanol. After the addition of 20µL internal standard solution (150 pg/µL each of [13C5]-FA, [13C5]-THF, [13C5]-5-MTHF, and [13C5]-5-FTHF) the samples were thoroughly mixed. Water (1 mL) containing 1% ascorbic acid and 1% methanol was added prior to purification using SPE columns.
Solid-phase extraction
Supelco LC-18 3 mL SPE cartridges were conditioned with 1 mL methanol, which was followed by 1 mL of SPE buffer. After loading the sample (1.5 mL), the cartridge was washed with SPE buffer (2 mL) followed by 0.5 mL eluting buffer (60% methanol and 0.2% ascorbic acid). Finally, 1 mL eluting buffer was used to elute folates from the cartridge. The eluate was evaporated under nitrogen and re-dissolved in eluting water (200 µL). After centrifugation for 5 min at 12000 rpm an aliquot (50 µL) was analyzed for THF, 5-MTHF, and FA by LC-MRM/MS.
Conversion of 5-FTHF and 10-FTHF into 5,10-MTHF
After analyzing THF, 5-MTHF, and FA, 1 MHCl (40µL) was added to each vial and the sample was kept at room temperature for 4h. This resulted in quantitative conversion of 5-FTHF and 10-FTHF into 5,10-MTHF and [13C5]-5-FTHF into [13C5]-5,10-MTHF. An aliquot of the resulting solution (50 µL) was then analyzed by LC-MRM/MS.
Validation study
The validation study was performed (n=5) on the QC samples. The lower limit of quantitation (LLOQ) QC samples were 4.4; 4.5, 4.4, 4.5 nmol/L for 5-MTHF, THF, 5,10-MTHF, and FA, respectively. The lower QC (LQC) samples were 10.9, 11.2, 11.0, 11.3 nmol/L for 5-MTHF, THF, 5,10-MTHF, and FA, respectively. The middle QC (MQC) samples were 43.5, 44.9, 43.8, 45.3 nmol/L for 5-MTHF, THF, 5,10-MTHF, and FA, respectively. The high QC (HQC) samples were 174.1, 179.6, 175.3, 181.2 nmol/L for 5-MTHF, THF, 5,10-MTHF, and FA, respectively. The upper QC (UQC) sample was 870.6 nmol/L for 5-MTHF only.
Replicate analysis of RBC folates
Whole blood and plasma folates from five subjects were each analyzed five times using the methods described above and RBC folate concentrations determined. A separate whole blood and plasma sample from one subject was analyzed in duplicate on 15 separate occasions over a 1-year period in order to determine the precision of the assay over time and the stability of samples stored at −80°C.
Data analysis
All data analysis was performed using Analyst software, version 1.41 (Concord, ON, Canada) from raw mass spectral data. Calibration curves were plotted using a linear regression with weighting index of 1/x. Concentrations of folates in validation samples were determined from the calibration line, and used to calculate the accuracy and precision of the method within the study.
Calculation of RBC folate concentrations
RBC folate concentrations were calculated according to the method of Lamers et al.:43
RESULTS AND DISCUSSION
Conversion of 5-FTHF and 10-FTHF into 5,10-MTHF
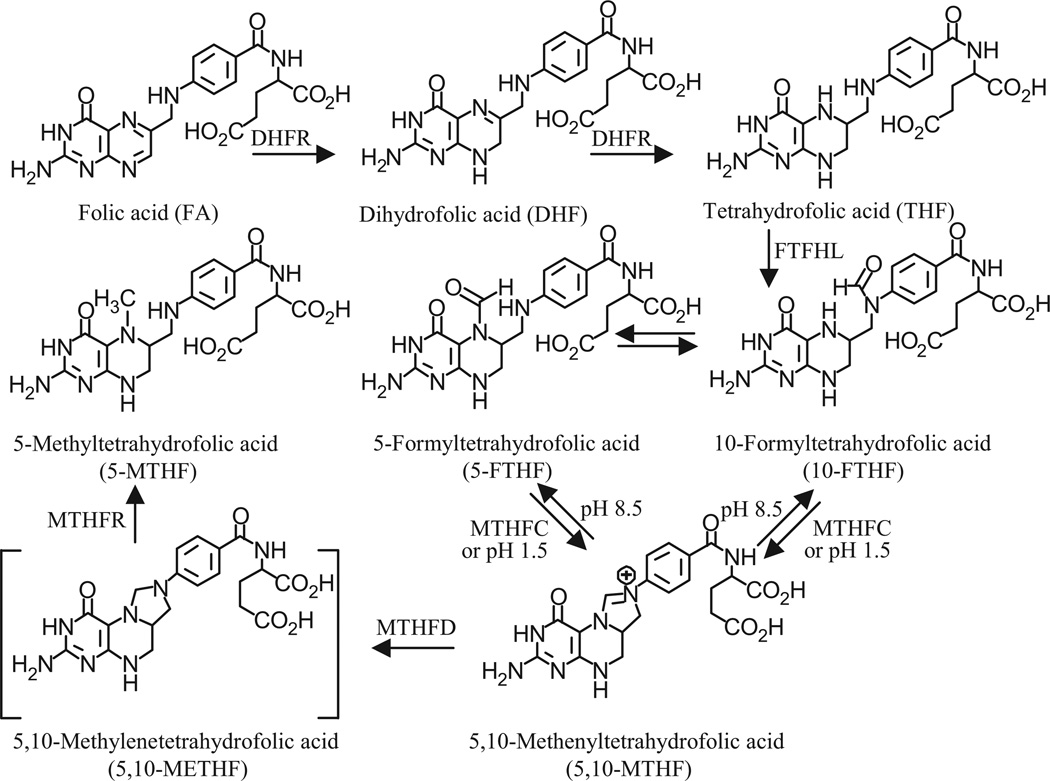
5-FTHF and 10-FTHF undergo dehydration to 5,10-MTHF under acidic conditions in a pH-dependent manner (Fig. 3). In contrast, under neutral or alkaline pH conditions, 5,10-MTHF is converted into 10-FTHF, which then slowly interconverts with 5-FTHF. These different forms of folate are present in RBCs primarily in polyglutamated forms. It is common practice to initiate hydrolysis via activation of human plasma pteroylpolyglutamate hydrolase by lysing the RBCs under acidic conditions (pH 5) in the presence of ascorbic acid.20,40 Therefore, there is always some conversion of 5-FTHF and 10-FTHF into 5,10-MTHF. The alternative use of rodent serum pteroylpolyglutamate hydrolases with isolated lysed RBCs still requires an acidic pH for optimal activity.44 This made it difficult to reliably quantify the individual amounts of 5-FTHF, 10-FTHF, and 5,10-MTHF in the RBCs. Conversion of 5-FTHF and 10-FTHF into 5,10-MTHF proceeded quantitatively at pH 1.5. As a result, it was possible to accurately quantify the pool of folate precursors available for conversion into 5,10-METHF simply by acidifying the RBC extract (Fig. 3). Unfortunately, under these conditions, THF was unstable. Therefore, samples were analyzed initially for FA, THF, and 5-MTHF. They were then acidified to pH 1.5 with 1M HCl ready for analysis of the 5,10-METHF precursors as 5,10-MTHF (Fig. 3). The [13C5]-5-FTHF internal standard was quantitatively converted into [13C5]-5,10-MTHF, which made it possible to accurately quantify the amount of unlabeled 5-FTHF and 10-FTHF, as well as any 5,10-MTHF that had been present in the sample originally. By monitoring the MRM channels for [13C5]-5-FTHF, as well as unlabeled 5-FTHF, and 10-FTHF, it was also possible to ensure that quantitative conversion into 5,10-MTHF had occurred (data not shown). 5,10-METHF is extremely unstable under both acidic and basic conditions and so it is not possible to analyze this form of folate in RBCs. However, it is so rapidly utilized in cellular processes by enzymes such as MTHFR, serine hydroxymethyltransferase, and thymidylate synthase (Fig. 1) that it is unlikely to be present in significant quantities.
Figure 3.
Biosynthesis and interconversion of folates. Abbreviations are as in Fig. 1
LC/MS
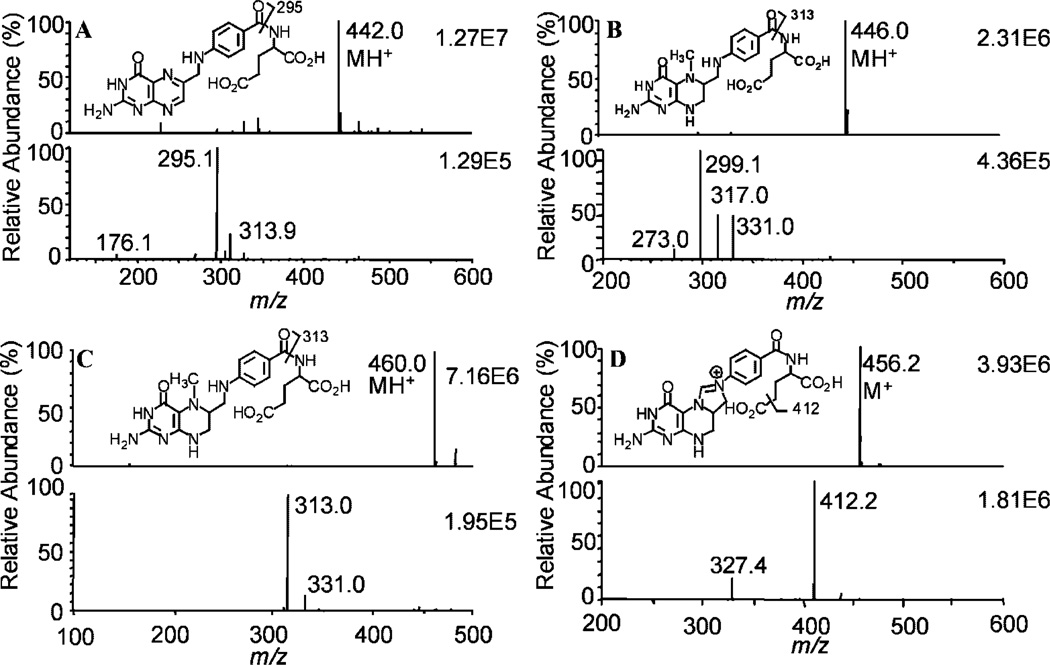
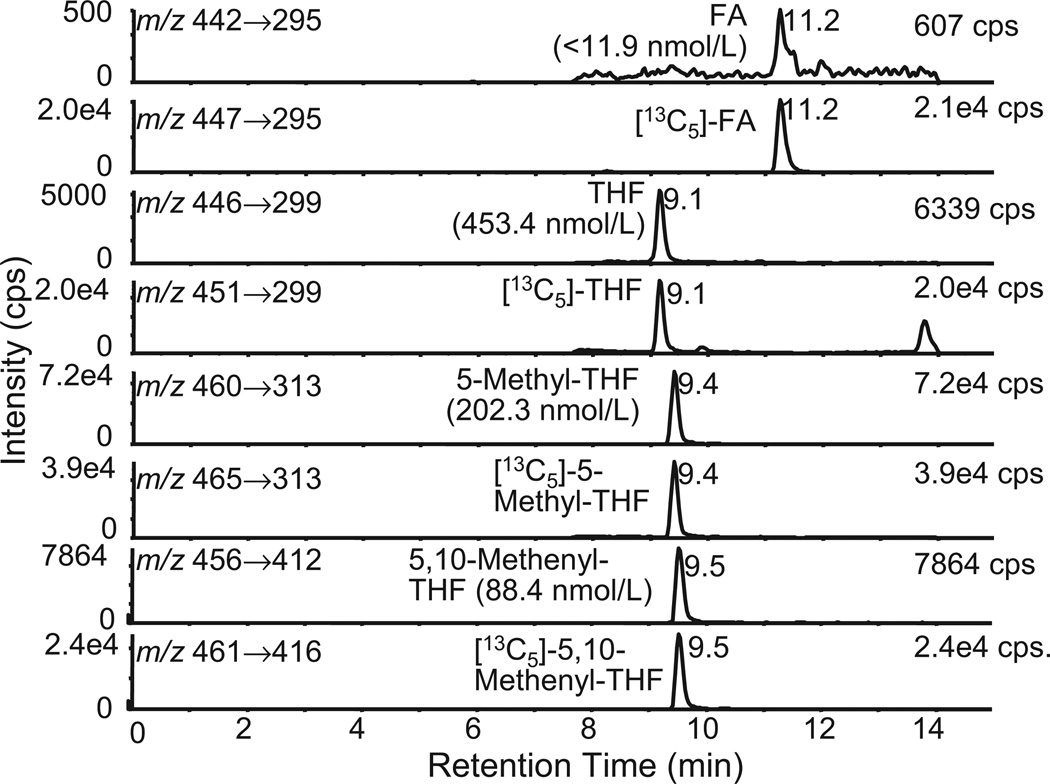
Under positive turboionspray conditions, the most abundant folate ions arose from the protonated molecules (MH+), except for 5,10-MTHF, which is already charged (M+). In the full scan mode, FA, THF, 5-MTHF, and 5,10-MTHF had precursor ions at m/z 442, 446, 460, and 456, respectively (Figs. 2(A)–2(D)). The most abundant product ions observed for FA, THF, and 5-MTHF after collision-induced dissociation and MS/MS analysis corresponded to loss of the γ-glutamate residue (Fig. 2(A)–2(C)). On the other hand, the product ion corresponding to loss of the carboxyl group was the major product ion observed for 5,10-MTHF (Fig. 2(D)). Formic acid, acetic acid, and trifluoroacetic acid were tested as mobile phase additives to improve chromatographic peak shape and MS signal. Acetic acid (1%) gave the highest ion intensities so it was chosen for the folate analyses. Several columns were also tested in order to improve the separation of individual folates from any endogenous interfering signals. Excellent separations were observed using the cyanopropyl column; however, substantial peak tailing occurred. In contrast, the YMC C18 AQ column separated each of the folates and gave excellent peak shapes (Figs. 4 and 5). Retention times were between 9 and 12 min.
Figure 2.
Full scan (upper panel) and product ion spectra (lower panel) of folates: (A) FA, (B) THF, (C) 5-MTHF, and (D) 5,10-MTHF.
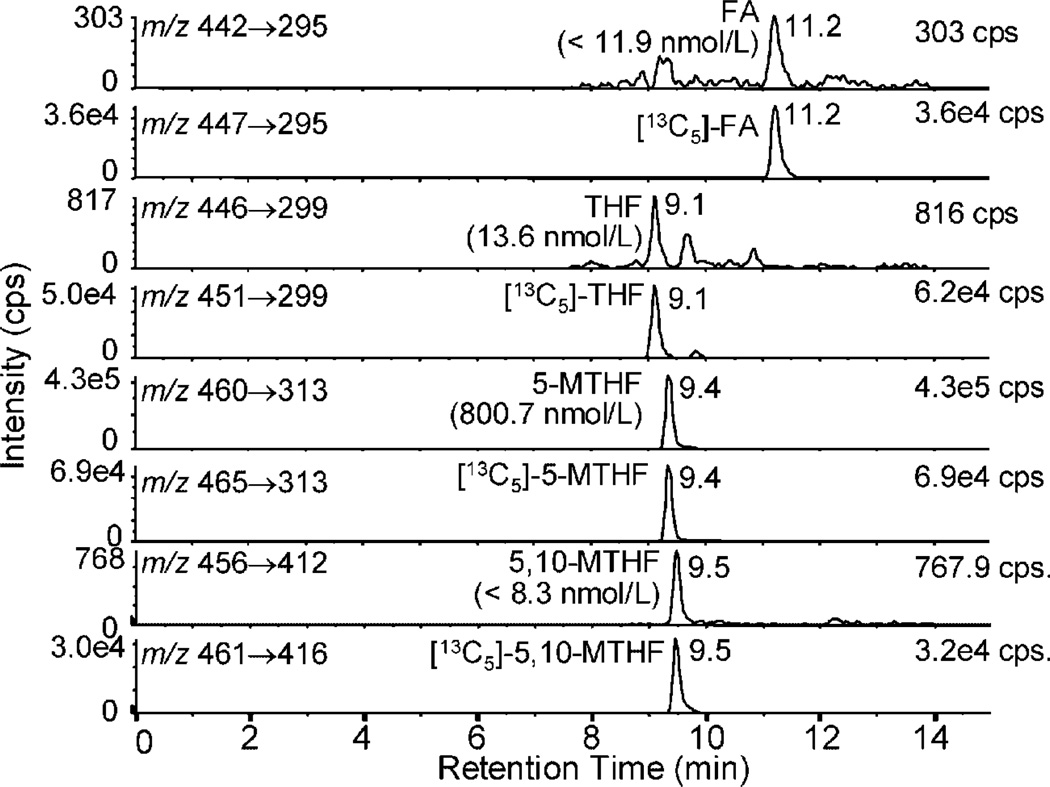
Figure 4.
LC-MRM/MS chromatograms of RBC folates from a homozygous MTHFR 677CC genotype that was a type I phenotype (5-MTHF >95%; THF <5%; 5,10-MTHF <5%).
Figure 5.
LC-MRM/MS chromatograms of RBC folates from a homozygous MTHFR 677TT genotype that was a type IV phenotype (5-MTHF <55%; THF >20%; 5,10-MTHF >5%).
Hydrolysis efficiency
Pteroylhepta-γ-L-glutamic acid was used to examine the hydrolysis efficiency of plasma pteroylpolyglutamate hydrolase in the pH range 4.0 to 7.0. As described previously,20 optimal hydrolysis occurred at pH 5 (data not shown). Replicate (n = 5) RBC preparations from five different subjects were then spiked with pteroylhepta-γ-L-glutamic acid, in order to monitor the efficiency of plasma human pteroylpolyglutamate hydrolase-mediated conversion of folylpolyglutamates into the corresponding monoglutamates. This approach was reported previously by Pfeiffer and Gregory for determining the hydrolysis efficiency of pteroylpolyglutamate hydrolase except that pteroylhepta-γ-L-glutamic acid was used instead of 5-MTHF-hepta-γ-L-glutamic acid.40 In our samples the hydrolysis efficiency was 98.7 ± 7.2% (n = 25).
Sensitivity and linearity
To determine the limit of detection (LOD), a serial dilution of folate was prepared (0.1 to 10 ng/mL). The LOD determined at a signal/noise (S/N) ratio of 3:1 for FA, THF, 5-MTHF, and 5,10-MTHF were 3,6,2.5, and 1.2 pg on-column, respectively. Sensitivities were similar to those reported recently.36 Calibration curves were prepared in the range of 4.5 to 900nmol/L. Samples were stored in 1% ascorbic acid containing 10mM 2-mercaptoethanol. Calibration curves for FA (y = 0.0109× − 0.0008; r2 0.9999), THF (y = 0.0070× − 0.0052; r2 0.9999), 5-MTHF (y = 0.0427× − 0.0652; r2 0.9996), and 5,10-MTHF (y = 0.0183× − 0.0442; r2 0.9973) were fitted to a linear regression with a 1/x weighting.
Accuracy and precision
Concentrations of folates in QC samples were determined from the calibration line on each occasion and are presented in Table 1 along with accuracy and precision values. At all QC sample concentrations examined, the accuracy was well within 100 ± 15% and the precision values were better than 15%. These criteria meet the guidelines on bioanalytical methods validation recommended by the FDA-sponsored meeting in Crystal City, VA in 2006.45 RBC samples from five individuals were each analyzed as five replicates, and again acceptable precision and accuracy were obtained (Table 2). Generally, 5-MTHF was the dominant form of folate in RBCs with much smaller amounts of THF and 5,10-MTHF (Table 2). Finally, an RBC sample from a single individual was analyzed in duplicate on 15 separate occasions over a 1-year period. 5-MTHF values were 1120.0 ± 46.6 nmol/L (coefficient of variance (CV) 4.2%), THF values were 26.2 ± 3.1 nmol/L (CV 11.8%), and 5,10-MTHF concentrations were 52.3 ± 6.0 nmol/L (CV 11.4%). Therefore, the assay was robust and long-term storage of the samples did not result in any deterioration of the individual RBC folates.
Table 1.
Precision and accuracy of folate analyses (n = 15)
| Analyte | Parameter | LLOQ (nmol/L) | LQC (nmol/L) | MQC (nmol/L) | HQC (nmol/L) | UQC (nmol/L) |
|---|---|---|---|---|---|---|
| FA | Mean | 4.8 | 11.3 | 44.6 | 179.7 | |
| Precision(%) | 8.7 | 2.7 | 3.0 | 2.9 | ||
| Accuracy(%) | 102.8 | 100.2 | 98.3 | 99.1 | ||
| 5-MTHF | Mean | 4.4 | 10.7 | 41.8 | 169.3 | 850.6 |
| Precision(%) | 4.1 | 3.5 | 5.2 | 3.7 | 3.3 | |
| Accuracy(%) | 98.4 | 97.5 | 96.1 | 97.3 | 97.7 | |
| THF | Mean | 4.7 | 11.4 | 44.7 | 178.5 | |
| Precision(%) | 13.9 | 8.5 | 5.9 | 5.0 | ||
| Accuracy(%) | 103.6 | 101.3 | 99.3 | 99.4 | ||
| 5,10-MTHF | Mean | 4.2 | 10.3 | 41.2 | 167.4 | |
| Precision(%) | 8.1 | 6.1 | 6.5 | 5.8 | ||
| Accuracy(%) | 96.9 | 94.7 | 94.1 | 95.5 |
Abbreviations: LLOQ, lower limit of quantitation; LQC, lower quality control, MQC, middle quality control; UQC, upper quality control; FA, folic acid; 5-MTHF, 5-methyltetrahydrofolate; THF, tetrahydrofolate; 5,10-MTHF, 5,10-methenyltetrahydrofolate.
Table 2.
Replicate analyses (n = 5) with different RBC samples (n = 5)
| Subject | FA (nmol/L) | CV | 5-MTHF (nmol/L) | CV | THF (nmol/L) | CV | 5,10-MTHF (nmol/L) | CV |
|---|---|---|---|---|---|---|---|---|
| A | ND | ND | 826.7 | 6.5% | 20.5 | 12.5% | ND | ND |
| B | ND | ND | 715.2 | 4.5% | 21.9 | 12.5% | ND | ND |
| C | ND | ND | 812.3 | 5.1% | 24.2 | 9.8% | ND | ND |
| D | ND | ND | 1269.4 | 3.9% | 60.9 | 7.4% | 17.5 | 7.1% |
| E | ND | ND | 738.4 | 2.0% | 32.0 | 14.9% | ND | ND |
| Mean | ND | ND | 826.7 | 4.4% | 31.9 | 11.4% | 3.5 | 7.8% |
Abbreviations: ND, not detected; other abbreviations as for Table 1.
Analysis of RBC folates
Mature RBCs are unable to accumulate or export folate derivatives,43,46 and the current folate content of each RBC reflects that present at the time of its formation through erythropoiesis.15 As the life span of a normal RBC is approximately 120 days, RBC folate measurements reflect the average levels during the preceding 4 months, in contrast to plasma or serum folate levels which exhibit transient fluctuations due in part to daily differences in dietary intake.47 Therefore, RBC folate content has been used as a surrogate biomarker for historical folate status over the medium term. The seminal finding that 5-FTHF was present in RBCs of individuals with the MTHFR 677TT genotype15 prompted the present study to develop methodology that could distinguish different phenotypes between and within the three MTHFR 677C>T genotype classes. This might then make it possible to further stratify disease risk associated with disruptions to the folate pathway. Stable isotope dilution LC-MRM/MS affords an opportunity to define such phenotypes with high precision. Although several groups have used similar methodology recently to quantify RBC folates,17,31,33,35,36,38 none have generated distinct phenotypes inclusive of the key folate metabolites. For example, Fazili and Pfeiffer have reported a high-throughput method in which 5-MTHF, THF, 5,10-MTHF, and 5-FTHF were analyzed directly in 38 subjects with defined MTHFR 677C>T genotypes.31 10-FTHF was unstable under the assay conditions used. In contrast to the present study, no distinct RBC folate phenotypes were reported for the three different MTHFR 677C>T genotypes. Furthermore, unexpectedly high 5-FTHF concentrations were found in MTHFR 677CC homozygotes compared with the previous study of Bagley and Selhub.15 It was suggested that this might have been due to the selective loss of low amounts of formylated folates from the CC samples during the extraction procedure.31 The study of Smulders et al. conducted on 104 subjects with defined MTHFR 677C>T genotypes simply compared methylated and non-methylated folates and did not distinguish between THF and 5,10-MTHF.17 This study showed that the MTHFR 677C>T genotype was the dominant determinant of non-methylfolate accumulation. Thus, the T-allele and total folate status were positively and independently correlated with non-methylfolate accumulation. Finally, Fazili et al.38 analyzed RBC folates in 171 MTHFR-genotyped blood bank samples using the high-throughput method in which 10-FTHF was unstable. This study confirmed the findings of Smulders et al.17 by showing that the TT genotypes had an accumulation of non-methylfolates in their whole blood samples.38
The analysis of RBC folates provides a significant bioanalytical challenge because the individual forms of folates are retained in the RBCs after erythropoiesis by virtue of polyglutamylation in which varying numbers of glutamate residues are added to the folates. In order to rigorously quantify the individual folates, it is necessary to first hydrolyze the folylpolyglutamates to their corresponding monoglutamate forms (Fig. 3). Plasma enzymes that typically perform this function have optimal activities at acidic pH, under which conditions formylated folates are converted into 5,10-MTHF. Therefore, we chose to analyze the formylated folate derivatives as 5,10-MTHF after their acid-catalyzed conversion to the latter. This conversion was conducted in the presence of [13C5]-5-FTHF to ensure that no residual formylated folate derivatives remained by including the relevant MRM transitions in the LC/MS analyses. The resulting [13C5]-5,10-MTHF was then used as the internal standard to quantify all of the unlabeled 5,10-MTHF that had been formed from 5-FTHF and 10-FTHF as well as any unlabeled 5,10-MTHF that had been present at the time of sample collection.
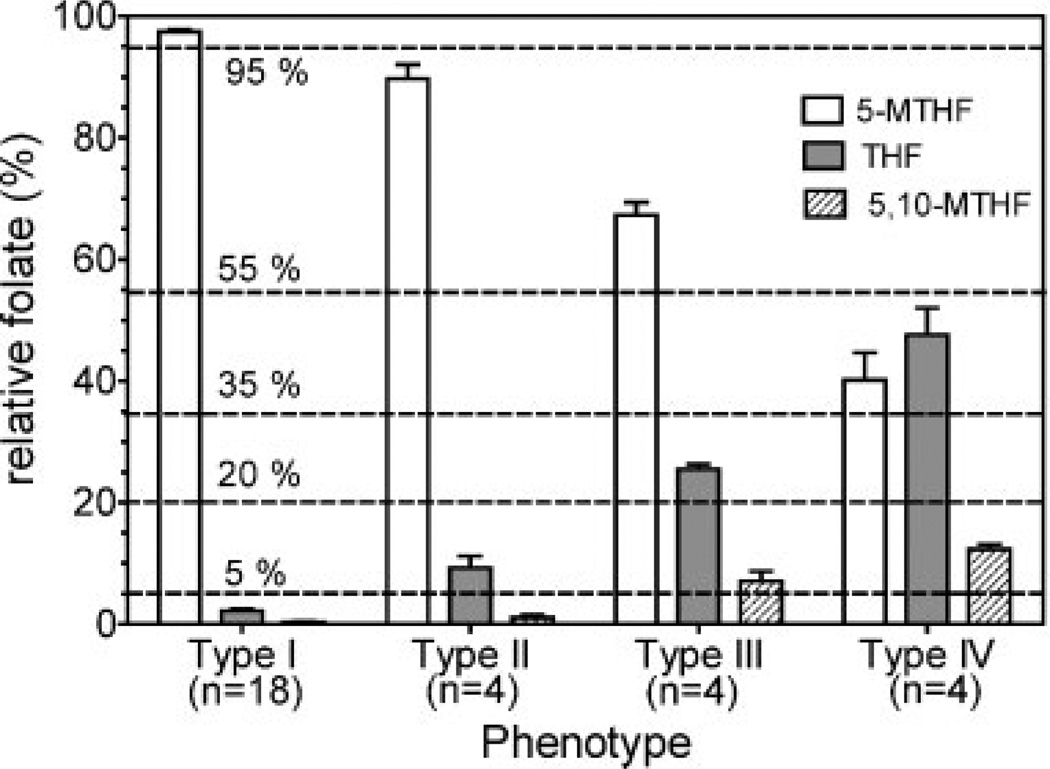
Analyses of RBC folates from 30 genotyped individuals (ten MTHFR 677CC homozygotes, ten MTHFR 677CT heterozygotes, and ten MTHFR 677TT homozygotes) were conducted using the stable isotope dilution LC-MRM/MS methodology. As expected, the dominant form of folate in most samples was 5-MTHF (Table 3). However, there were distinct differences in folate distribution patterns between and within the MTHFR 677C>T genotype classes. Generally, RBCs from both MTHFR 677 CC homozygotes and CT heterozygotes had very low levels of THF (i.e. <5% total folate) and almost undetectable amounts of 5,10-MTHF (Fig. 4; Table 3). However, THF, but not 5,10-MTHF, could be detected at a higher level (i.e. >5% total folate) in three of the CT heterozygotes. This resulted in a higher mean THF value of 42.0 nmol/L or 4.3% of total folates for the ten CT genotypes that were analyzed (Table 3). The above two folate distribution patterns suggested the existence of two distinct phenotypes, designated type I (5-MTHF >95%; THF <5%; 5,10-MTHF <5%) and type II (5-MTHF <95%; THF 5% to 20%; 5,10-MTHF <5%), respectively, within the two main genotype classes (Fig. 6). In contrast to the CC and CT genotypes, 5,10-MTHF was readily detectable (mean 8.0% of total folate) in RBCs from TT homozygotes (Table 3) indicating that the TT phenotypes are more complex than the CC and CT genotypes with respect to RBC folate distribution. Furthermore, RBCs from TT homozygotes had much higher amounts of THF (mean 30.2% of total folate) than RBCs from those with the CC and CT genotypes (Table 3). There appears to be at least two distinct TT phenotypes, defined by the relative amounts of 5-MTHF, THF, and 5,10-MTHF (Fig. 6). Accordingly, the two phenotypes within the MTHFR 677TT genotype class have been designated type III (5-MTHF >55%; THF >20%; 5,10-MTHF >5%) and type IV (5-MTHF <55%; THF >20%; 5,10-MTHF >5%), as shown in Fig. 6. Pending further investigation involving larger numbers of individuals, the type III and IV phenotypes can be collectively identified by THF and 5,10-MTHF levels that exceed 20% and 5% of total RBC folate, with further subdivision being defined by 5-MTHF concentrations above 55% or below 55% of total folates, respectively (Fig. 6). Intriguingly, one of the ten TT homozygotes had a type I phenotype and another had a type II phenotype. Thus, MTHFR 677TT genotype alone does not appear to be sufficient unequivocally to define the phenotype of a particular subject, suggesting that, for a minority of MTHFR 677TT homozygotes, additional biochemical and/or genetic variables are involved in determining the relative amounts of individual folate metabolites.
Table 3.
Mean folate content of RBCs (n = 30) from three different MTHFR 677C>T genotypes
| Subject | 5-MTHF |
THF |
5,10-MTHF |
Total nmol/L |
|||
|---|---|---|---|---|---|---|---|
| nmol/L | % | nmol/L | % | nmol/L | % | ||
| CC mean (n = 10) | 929.6 | 97.7 | 21.3 | 2.0 | 4.0 | 0.4 | 955.0 |
| SD | 285.7 | — | 15.3 | — | 6.5 | — | — |
| CT mean (n = 10) | 1065.4 | 95.2 | 42.0 | 4.3 | 5.4 | 0.5 | 1112.8 |
| SD | 360.4 | — | 34.7 | — | 7.4 | — | — |
| TT mean (n = 10) | 764.0 | 61.8 | 374.5 | 30.2 | 100.4 | 8.0 | 1239.0 |
| SD | 292.3 | — | 249.4 | — | 73.5 | — | — |
Figure 6.
Folate phenotypes based on the relative amounts of RBC folates: type I (5-MTHF >95%; THF <5%; 5,10-MTHF <5%) comprised ten CC, seven CT, and one TT genotype; type II (5-MTHF <95%; THF >5% to 20%; 5,10-MTHF <5%) comprised three CT and one TT genotype; type III (5-MTHF >55%; THF >20%; 5,10-MTHF >5%) comprised four TT genotypes; type IV (5-MTHF <55%; THF >20%; 5,10-MTHF >5%) comprised four TT genotypes.
The finding of increased THF and 5,10-MTHF concentrations in RBCs from MTHFR 677TT homozygotes is in keeping with the concept that the thermolabile enzyme variant defined by this genotype has impaired capacity for mediating the conversion of 5,10-METHF into 5-MTHF. Alternative biochemical pathways such as those involved in DNA synthesis can then be up-regulated, in utilizing the 5,10-METHF that is not converted into 5-MTHF, and thereby favoring the accumulation of THF and 5,10-MTHF. This can occur through increased thymidylate synthase-mediated thymidine phosphate biosynthesis (with concomitant formation of THF) as well as from 10-FTHF-mediated increases in purine biosynthesis, which result from conversion of 5,10-METHF into 10-FTHF by MTHFD, a trifunctional enzyme with both dehydrogenase and cyclohydrolase activity (Fig. 1).48
CONCLUSIONS
The analysis of RBC folates provides a surrogate biomarker for folate status of an individual because RBCs are unable to transport and accumulate folate derivatives.47 Therefore, they reflect the folate status of an individual at the time of erythropoiesis.15 Individual folates are present as folylpolyglutamates, which prevents their secretion from the RBCs. Therefore, it was first necessary to convert them into the corresponding monoglutamates prior to analysis. This was accomplished using an individual’s own plasma pteroylpolyglutamate hydrolase by simply lysing the RBCs in a whole blood sample. Optimal polyglutamate hydrolysis occurs at pH 5 so the whole blood was treated with ascorbic acid in order to reduce the pH to this level.20,40 The ascorbic acid also served as an antioxidant to prevent loss of the labile THF derivatives. Unfortunately, under these conditions, substantial amounts of the formylated THFs were dehydrated to 5,10-MTHF. Therefore, the dehydrations were allowed to go to completion at pH 1.5 in the presence of [13C5]-5-FTHF, which was also converted into [13C5]-5,10-MTHF. The resulting [13C5]-5,10-MTHF was then used as an internal standard for the formylated forms of THF that had been converted into 5,10-MTHF as well as the 5,10-MTHF that was present in the original sample (Fig. 3). A stable isotope dilution LC-MRM/MS method was developed for the accurate and precise quantification of the spectrum of the resulting RBC folates. The method was sensitive and robust, and was used to assess the relationship between different MTHFR 677C>T genotypes and RBC folates in 30 genotyped subjects. This indicated that there are four different phenotypes that are differentially distributed between the MTHFR 677C>T genotype classes (Fig. 6). The assay is now being employed in combination with analyses of homocysteine4 and glutathione11 for extensive phenotyping studies in human populations with defined genotypes.
The methodology described here has the potential to identify subgroups of individuals with genotype/phenotype profiles that confer excess risk of pathologies that are known to be associated with dysfunction in folate/homocysteine metabolism. Such genotype/phenotype-based risk estimation may in the future be used in the conduct of clinical studies and to develop predictive and diagnostic screening protocols.
Acknowledgements
We acknowledge the support of grants RO1AR47663, RO1CA108862, UL1RR024134, and P30ES0135080 from the National Institutes of Health, and grant PA4100038714 from the Commonwealth of Pennsylvania Department of Health. We thank Dr. Rudolf Moser of Merck Eprova AG, Schaffhausen, Switzerland, for the kind gift of folate standards.
REFERENCES
- 1.Selhub J. J Nutr. Health Aging. 2002;6:39. [PubMed] [Google Scholar]
- 2.Stover PJ. Nutr. Rev. 2004;62:S3. doi: 10.1111/j.1753-4887.2004.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 3.Smulders YM, Stehouwer CD. Semin. Vasc. Med. 2005;5:87. doi: 10.1055/s-2005-872395. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Lu ZY, Brown KS, Whitehead AS, Blair IA. Biomed. Chromatogr. 2007;21:107. doi: 10.1002/bmc.735. [DOI] [PubMed] [Google Scholar]
- 5.Brown KS, Huang Y, Lu ZY, Jian W, Blair IA, Whitehead AS. Atherosclerosis. 2006;189:133. doi: 10.1016/j.atherosclerosis.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Pitkin RM. Am. J. Clin. Nutr. 2007;85:285S. doi: 10.1093/ajcn/85.1.285S. [DOI] [PubMed] [Google Scholar]
- 7.Jensen LE, Etheredge AJ, Brown KS, Mitchell LE, Whitehead AS. Am. J. Med. Genet. A. 2006;140:1114. doi: 10.1002/ajmg.a.31212. [DOI] [PubMed] [Google Scholar]
- 8.Kim YI. Mol. Nutr. Food Res. 2007;51:267. doi: 10.1002/mnfr.200600191. [DOI] [PubMed] [Google Scholar]
- 9.Sanderson P, Stone E, Kim YI, Mathers JC, Kampman E, Downes CS, Muir KR, Baron JA. Br. J. Nutr. 2007;98:1299. doi: 10.1017/S0007114507771908. [DOI] [PubMed] [Google Scholar]
- 10.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Proc. Natl. Acad. Sci. USA. 1997;94:3290. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu P, Oe T, Blair IA. Rapid Commun. Mass Spectrom. 2008;22:432. doi: 10.1002/rcm.3380. [DOI] [PubMed] [Google Scholar]
- 12.Schwahn B, Rozen R. Am. J. Pharmacogenomics. 2001;1:189. doi: 10.2165/00129785-200101030-00004. [DOI] [PubMed] [Google Scholar]
- 13.Pejchal R, Campbell E, Guenther BD, Lennon BW, Matthews RG, Ludwig ML. Biochemistry. 2006;45:4808. doi: 10.1021/bi052294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strain JJ, Dowey L, Ward M, Pentieva K, McNulty H. Proc. Nutr. Soc. 2004;63:597. doi: 10.1079/pns2004390. [DOI] [PubMed] [Google Scholar]
- 15.Bagley PJ, Selhub J. Proc. Natl. Acad. Sci. USA. 1998;95:13217. doi: 10.1073/pnas.95.22.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis SR, Quinlivan EP, Shelnutt KP, Maneval DR, Ghandour H, Capdevila A, Coats BS, Wagner C, Selhub J, Bailey LB, Shuster JJ, Stacpoole PW, Gregory JF., III J. Nutr. 2005;135:1040. doi: 10.1093/jn/135.5.1040. [DOI] [PubMed] [Google Scholar]
- 17.Smulders YM, Smith DE, Kok RM, Teerlink T, Gellekink H, Vaes WH, Stehouwer CD, Jakobs C. J.Nutr. Biochem. 2007;18:693. doi: 10.1016/j.jnutbio.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Botto LD, Yang Q. Am. J. Epidemiol. 2000;151:862. doi: 10.1093/oxfordjournals.aje.a010290. [DOI] [PubMed] [Google Scholar]
- 19.Lewis SJ, Ebrahim S, Davey SG. Br. Med. J. 2005;331:1053. [Google Scholar]
- 20.Quinlivan EP, Hanson AD, Gregory JF. Anal. Biochem. 2006;348:163. doi: 10.1016/j.ab.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Molloy AM, Scott JM. Methods Enzymol. 1997;281:43. doi: 10.1016/s0076-6879(97)81007-5. [DOI] [PubMed] [Google Scholar]
- 22.Molloy AM, Daly S, Mills JL, Kirke PN, Whitehead AS, Ramsbottom D, Conley MR, Weir DG, Scott JM. Lancet. 1997;349:1591. doi: 10.1016/S0140-6736(96)12049-3. [DOI] [PubMed] [Google Scholar]
- 23.McGown EL, Lewis CM, Dong MH, Sauberlich HE. Clin. Chem. 1978;24:2186. [PubMed] [Google Scholar]
- 24.Lucock MD, Hartley R, Smithells RW. Biomed. Chromatogr. 1989;3:58. doi: 10.1002/bmc.1130030204. [DOI] [PubMed] [Google Scholar]
- 25.Bagley PJ, Selhub J. Clin. Chem. 2000;46:404. [PubMed] [Google Scholar]
- 26.Freisleben A, Schieberle P, Rychlik M. Anal. Biochem. 2003;315:247. doi: 10.1016/s0003-2697(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 27.Santhosh-Kumar CR, Deutsch JC, Hassell KL, Kolhouse NM, Kolhouse JF. Anal. Biochem. 1995;225:1. doi: 10.1006/abio.1995.1099. [DOI] [PubMed] [Google Scholar]
- 28.Santhosh-Kumar CR, Deutsch JC, Ryder JW, Kolhouse JF. Eur. J. Clin. Nutr. 1997;51:188. doi: 10.1038/sj.ejcn.1600385. [DOI] [PubMed] [Google Scholar]
- 29.Garbis SD, Melse-Boonstra A, West CE, van Breemen RB. Anal. Chem. 2001;73:5358. doi: 10.1021/ac010741y. [DOI] [PubMed] [Google Scholar]
- 30.Pfeiffer CM, Fazili Z, McCoy L, Zhang M, Gunter EW. Clin. Chem. 2004;50:423. doi: 10.1373/clinchem.2003.026955. [DOI] [PubMed] [Google Scholar]
- 31.Fazili Z, Pfeiffer CM. Clin. Chem. 2004;50:2378. doi: 10.1373/clinchem.2004.036541. [DOI] [PubMed] [Google Scholar]
- 32.Owens JE, Holstege DM, Clifford AJ. J. Agric. Food Chem. 2005;53:7390. doi: 10.1021/jf0510485. [DOI] [PubMed] [Google Scholar]
- 33.Fazili Z, Pfeiffer CM, Zhang M, Jain R. Clin. Chem. 2005;51:2318. doi: 10.1373/clinchem.2005.053801. [DOI] [PubMed] [Google Scholar]
- 34.Nelson BC, Satterfield MB, Sniegoski LT, Welch MJ. Anal. Chem. 2005;77:3586. doi: 10.1021/ac050235z. [DOI] [PubMed] [Google Scholar]
- 35.Satterfield MB, Sniegoski LT, Sharpless KE, Welch MJ, Hornikova A, Zhang NF, Pfeiffer CM, Fazili Z, Zhang M, Nelson BC. Anal. Bioanal. Chem. 2006;385:612. doi: 10.1007/s00216-006-0434-1. [DOI] [PubMed] [Google Scholar]
- 36.Smith DE, Kok RM, Teerlink T, Jakobs C, Smulders YM. Clin. Chem. Lab Med. 2006;44:450. doi: 10.1515/CCLM.2006.085. [DOI] [PubMed] [Google Scholar]
- 37.Owens JE, Holstege DM, Clifford AJ. J. Agric. Food Chem. 2007;55:3292. doi: 10.1021/jf063648p. [DOI] [PubMed] [Google Scholar]
- 38.Fazili Z, Pfeiffer CM, Zhang M, Jain RB, Koontz D. Clin. Chem. 2008;54:197. doi: 10.1373/clinchem.2007.096545. [DOI] [PubMed] [Google Scholar]
- 39.Ueland PM, Midttun O, Windelberg A, Svardal A, Skalevik R, Hustad S. Clin. Chem. Lab. Med. 2007;45:1737. doi: 10.1515/CCLM.2007.339. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer CM, Gregory JFIII. Clin. Chem. 1996;42:1847. [PubMed] [Google Scholar]
- 41.Available: http://snp500cancer.nci.nih.gov
- 42.Blakely RL. The biochemistry of folic acid and related pteridines. In: Neuberger A, Tatum EL, editors. Frontiers of Biology. Vol. 92 London: North Holland; 1969. [Google Scholar]
- 43.Lamers Y, Prinz-Langenohl R, Bramswig S, Pietrzik K. Am. J. Clin. Nutr. 2006;84:156. doi: 10.1093/ajcn/84.1.156. [DOI] [PubMed] [Google Scholar]
- 44.Thomas PM, Flanagan VP, Pawlosky RJ. J. Agric. Food Chem. 2003;51:1293. doi: 10.1021/jf020902e. [DOI] [PubMed] [Google Scholar]
- 45.Chaudhary AJ, Wickremsinhe ER, Berna MJ, Ackermann BL. Am. Drug Discov. 2006;1:34. [Google Scholar]
- 46.Bailey LB. J. Nutr. 1990;120(Suppl 11):1508. doi: 10.1093/jn/120.suppl_11.1508. [DOI] [PubMed] [Google Scholar]
- 47.Herbert V. Am. J. Hematol. 1987;26:199. doi: 10.1002/ajh.2830260211. [DOI] [PubMed] [Google Scholar]
- 48.Prasannan P, Pike S, Peng K, Shane B, Appling DR. J. Biol. Chem. 2003;278:43178. doi: 10.1074/jbc.M304319200. [DOI] [PMC free article] [PubMed] [Google Scholar]