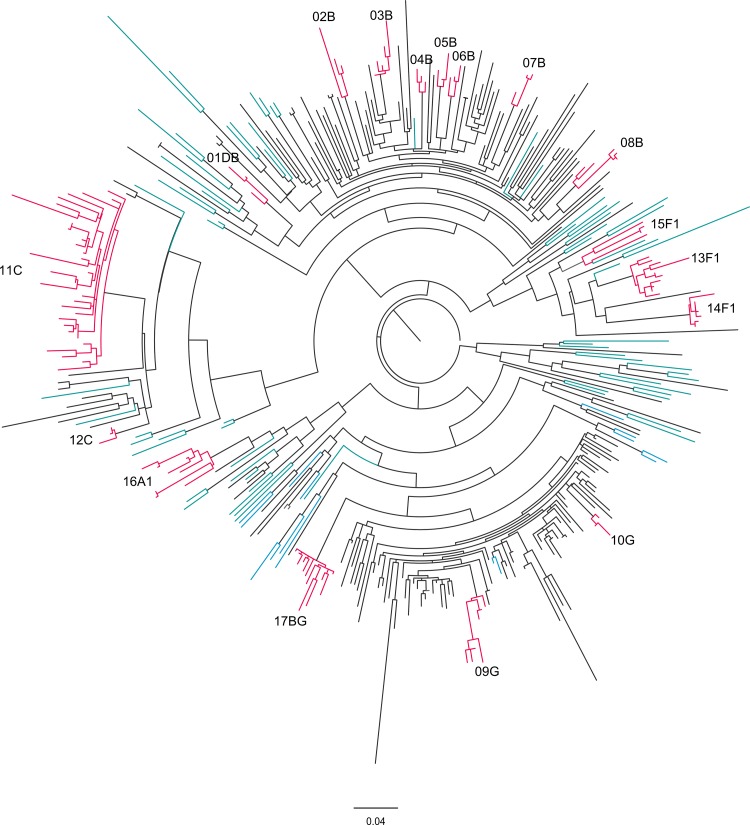
FIG 2.
Phylogenetic relationships among the HIV-1 sequences isolated from 289 infected individuals from Minho province, Portugal. Maximum likelihood (ML) phylogenetic analysis was performed using 289 partial HIV-1 sequences obtained in this study and 89 subtype reference sequences (colored in blue) and rooted using the M-group consensus sequence. The branch lengths are expressed as the number of nucleotide substitutions per site. The transmission clusters (colored in pink) were supported by an ML bootstrap support of ≥95% based on 1,000 replicates, an average genetic distance of ≤0.03 substitutions per site, and a Bayesian posterior probability equal to 1.