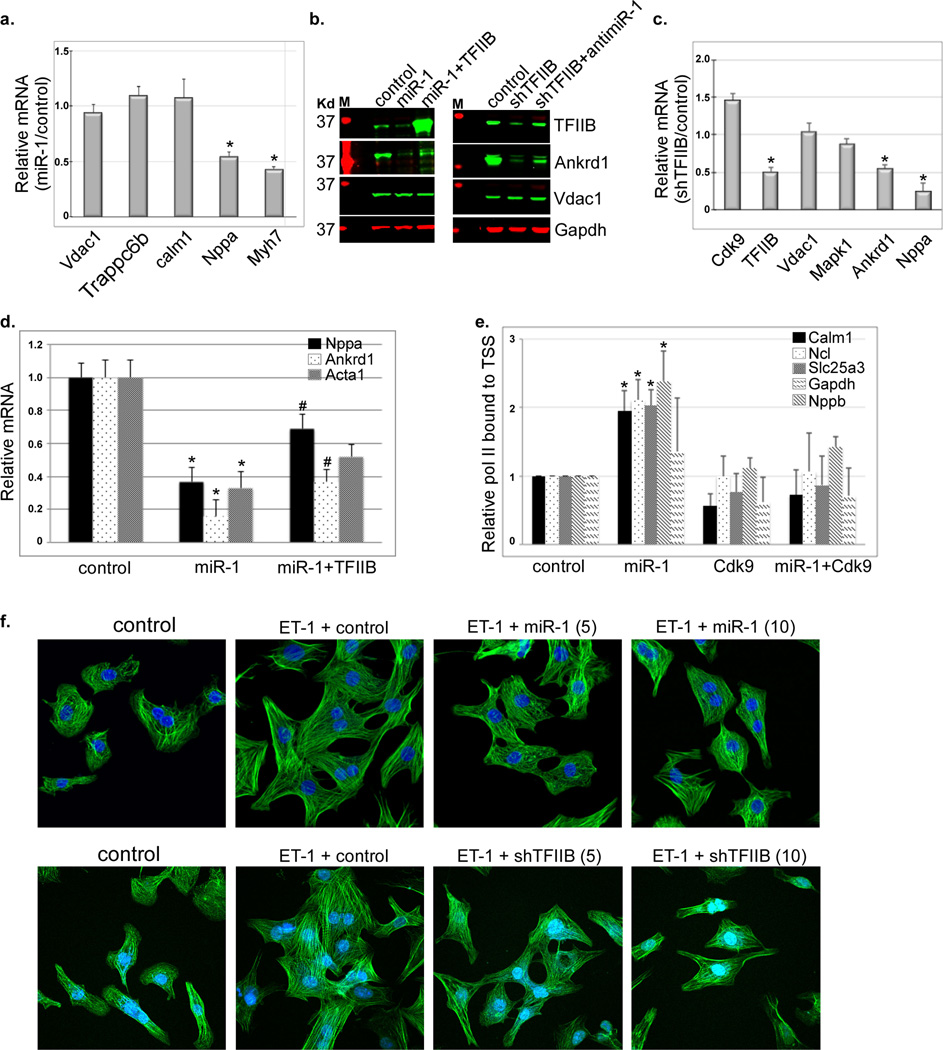
Figure 4. Knockdown of TFIIB selectively inhibits genes regulated by de novo recruitment of pol II and inhibits myocyte hypertrophy.
a. RNA extracted from neonatal myocytes supplemented with a control or Ad.miR-1 for 24 h was analyzed by qPCR. The data for 5 genes are averaged and plotted as relative miR-1/control values (n=3). Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control. b. Protein was extracted from cells treated as in (a.) in the absence or presence of exogenous Ad.TFIIB, or with a shRNA targeting TFIIB (Ad.shTFIIB) in the absence or presence of Ad.antimiR-1, and analyzed by Western blotting. c. RNA extracted from neonatal myocytes supplemented with a control or Ad.shTFIIB for 24 h was analyzed by qPCR (n=3). The data for 6 genes are averaged and plotted as relative shTFIIB/control values. Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control. d. RNA was extracted from cells treated as in (a.) in the absence or presence of exogenous Ad.TFIIB, and analyzed by qPCR for the specified genes (n=3). e. Myocytes were treated with control, Ad.miR-1, Ad.Cdk9, or Ad.miR-1+Cdk9 for 24. Cells were then fixed and subjected to ChIP-qPCR encompassing the TSS of the specified genes. The results were averaged and plotted as relative bound pol II. * is p < 0.05 v. control. f. Myocytes were treated with control (10 moi), Ad.miR-1 (5 or 10 moi, upper pannels), or Ad.shTFIIB (5 or 10 moi, lower panels) for 24 h before stimulating them with 100 nM endothelin (ET-1). After 24 h the cells were fixed and stained with Dapi and phalloidin.